Virtual Library
Start Your Search
Lluis Esteban Tejero
Moderator of
-
+
OA06 - Refining Lung Cancer Screening (ID 131)
- Event: WCLC 2019
- Type: Oral Session
- Track: Screening and Early Detection
- Presentations: 8
- Now Available
- Moderators:Tomasz Grodzki, Lluis Esteban Tejero
- Coordinates: 9/09/2019, 11:00 - 12:30, Hilton Head (1978)
-
+
OA06.01 - Comparison Between Radiomics-Based Machine Learning and Deep Learning Image Classification for Sub-Cm Lung Nodules (Now Available) (ID 2810)
11:00 - 12:30 | Presenting Author(s): Calum Macaulay | Author(s): Ian Janzen, Saeed Seyyedi, Ronald Abraham, Sukhinder Kaur Atkar-Khattra, John Mayo, Ren Yuan, Renelle Myers, Stephen Lam
- Abstract
- Presentation
Background
New clinical challenges have arisen from the recent recognition for an improved mortality of cancers via lung cancer screening using LDCT. A particular challenge for physicians and CADx systems is the classification and prediction of behavior for sub-cm lung nodules that are frequently present in screening CT scans. By predicting and classifying the behavior of these small nodules, we can identify potential cancerous nodules into the earlier stages of malignancy making them more easily treatable.
Method
We have evaluated a multitude of image processing techniques to assist in CADx systems for these small nodules such as Radiomic feature-based machine learning algorithms (linear discriminant analysis) as well as leveraging pretrained convolution neural networks such as VGG19 and InceptionV3 using deep learning/transfer learning techniques. The linear discriminate Radiomic analysis (LDA) classified a sample of CT imaged nodules (n=514) using quasi-volumetric nodule data (images of the nodules from CT slices above and below the central slice) into three discriminate categories: cancerous (clinically confirmed, n = 140) versus resolved (not present in follow up CT scans, n=107) versus stable (a negligible change in shape, texture, size in multi year follow up CT scans, n=267). Each nodule was segmented from the original CT scan using an inhouse lung CT image segmenation routine. This routine generated 2167 discrete CT nodule images upon which 133 Radiomic shape and texture features were calculated.
Result
The LDA Radiomic analysis correctly classified the individual nodual sections with an accuracy of 75.1% (jackknife - leave one out result) using only 18 features predefined traditional image analysis features (4 shape feature(s), 14 texture feature(s)) for cancer vs resolved + stable nodules. Requiring that more than or equal to 50% of sections from a nodule be classified as cancer for the nodule to be classified as cancer individual nodules could be correctly classified with an 80% accuracy.
The leveraged pretrained networks (VGG19, and InceptionV3) trained using standard data augmentation and finetuning techniques, trained on this same quasi-volumetric image data as a binary classification task (malignant vs. benign nodules) achieved an average classification accuracy of 71% and 75% respectively through 10-crossfold validation.
Conclusion
Machine learning using 18 Radiomics features was able to classify 75.1% of the 2167 CT nodule images (up to 5 images/CT slices per nodule) and 80% of the nodules correctly. The best of the Deep Learning networks achieved almost equivalent results.
The image classification deep neural network results suggest the implementation of more advanced regularization and initialization deep learning techniques to further refine the decision boundaries for these pretrained networks might be benefitial. We believe the development of visualization neural network software to highlight the defining nodule features during classification would clinically assist in providing context clues for nodule diagnosis.
This work has been supported by TFRI project ref:1068
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.02 - The Role of Simulation Modeling in Shaping Lung Cancer Screening Policies in the US and Elsewhere (Now Available) (ID 1012)
11:00 - 12:30 | Presenting Author(s): Rafael Meza
- Abstract
- Presentation
Background
Models of the natural history of cancer have played an important role in shaping cancer prevention and control policies across the world. Notably, the US National Cancer institute (NCI) Cancer Intervention and Surveillance Modeling Network (CISNET) consortium has developed multiple such models and a modeling infrastructure that has supported the development of guidelines and policies for cancer screening and tobacco control in the US and elsewhere.
Method
The CISNET Lung models incorporate and synthetize smoking and lung cancer data from clinical trials, epidemiological studies and surveillance systems. These models have informed US screening guidelines. But important questions remain as screening programs are being implemented, such as the relative effectiveness of risk-based versus pack-year eligibility strategies or the potential of cessation programs within the context of lung screening.
Result
Simulation of the US 1950 and 1960 birth-cohorts show that for a given number of screens, risk-based screening programs lead in general to higher mortality reductions than pack-year based strategies. This is also true for LYG, but the difference is less pronounced. Independently of the program, adding cessation interventions at the point of screening leads to considerable gains in LYG, and to a lesser effect on deaths prevented. E.g., under current guidelines and a 40% screening uptake scenario, adding a cessation intervention at the time of first screen with a 15% success probability, could increase LYG by 140% and lung cancer deaths prevented by 28% (fig). But the actual gains would greatly depend on coverage and the cessation probability (fig).
Conclusion
Simulation modeling provides a framework to extrapolate findings from clinical trials and epidemiological studies into population outcomes. This has shown to be key to be able to refine and identify lung cancer prevention optimal strategies for a given setting. And to gather support among stakeholders to adopt and implement such strategies.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.03 - An Open Source Lung Screening Management System (Now Available) (ID 1467)
11:00 - 12:30 | Presenting Author(s): Claudia I Henschke | Author(s): Anthony P Reeves, Ricardo Avila, Drew Moghanaki, Artit Jirapatnakul, David F. Yankelevitz
- Abstract
- Presentation
Background
Starting in 1992, the Early Lung Cancer Screening Project (ELCAP) investigators developed the ELCAP Management System (MS) to ensure high quality care and follow-up of the first 1,000 ELCAP participants. The resulting Lancet publication in 1999 created worldwide interest in screening and an updated web-based ELCAP MS was updated to be web-based and provided free of charge to participating institutions, together with the I-ELCAP protocol.
Method
The ELCAP MS was designed to be comprehensive and rapidly capture information on each participant to be used by coordinators, navigators, nurses, radiologists, and other medical professionals to ensure appropriate follow-up and care. It provides rapid documentation of telephone or other inquiries, registering, scheduling screening appointments, reporting results, diagnosis of lung cancer, and treatment, and archives all CT images for integrated access of image and patient information. It has been iteratively updated through user feedback, and supports medical reimbursement requirements and continuous quality improvement to minimize harms of lung screening across International ELCAP (I-ELCAP) sites.
Result
More than 81,000 participants in 80 institutions worldwide have contributed their LDCT findings and images. The MS has provided efficient data collection for rigorous assessment of screening outcomes which has resulted in some 300 publications and abstracts for protocol updating, comparisons, and continuous quality improvement.
Having anticipated “open science”, the ELCAP MS has been translated into an open source MS that offers a reference standard for data elements (1,500 data fields, 267 required) for robust and efficient management of lung screening programs. This first open source translation has been adopted by the United States Veterans Administration (VA) and integrated into its VistA Electronic Healthcare System for deployment at 10 VA medical centers through a grant for VA Partnership to increase Access to Lung Screening (VA-PALS). The software is being certified by the Open Source Electronic Health Record Alliance (OSEHRA); source code is available on GitHub.
Automated quantitative tools have been developed for identification and characterization of nodules, emphysema, major airways, calcification scoring of coronary arteries, aortic valve, thoracic aorta, breast tissue, liver, bone, and image quality. These tools are integrated into the ELCAP MS, and in the future will provide automatically-generated quantitative LDCT reports.
Conclusion
The ELCAP MS and I-ELCAP protocol have helped define current global standards for lung screening. Its developers have now made the ELCAP MS publicly available through OSEHRA for support of lung screening programs of any scale throughout the world.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.04 - Discussant - OA06.01, OA06.02, OA06.03 (Now Available) (ID 3756)
11:00 - 12:30 | Presenting Author(s): Betty C Tong
- Abstract
- Presentation
Abstract not provided
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.05 - Evaluation of a Deep Learning-Based Automatic Classifier for the Classification of Perifissural Nodules (Now Available) (ID 1928)
11:00 - 12:30 | Presenting Author(s): Daiwei Han | Author(s): Marjolein A. Heuvelmans, Mieneke Rook, Monique D. Dorrius, Luutsen Van Houten, Noah Waterfield Price, Lyndsey C Pickup, Petr Novotny, Jerome Declerck, Matthijs Oudkerk, P.M.A Van Ooijen, Fergus Gleeson, Rozemarijn Vliegenthart
- Abstract
- Presentation
Background
Perifissural nodules (PFNs) comprise approximately 20% of screening-detected nodules and are almost certainly benign. Automatic PFN classification could therefore reduce the number of follow-up procedures required for nodule work-up. Prior work has shown some success in AI classification with limited datasets. Here we evaluate the performance of a new deep convolutional neural network (CNN) for PFN classification, trained on a dataset of nodules retrospectively collected from multiple European centers, including validation on an independent reader-study dataset.
Method
Data (1103 Patients, 1557 unique nodules and 3320 nodule images) were collected from three centers in the UK and the Netherlands. Each nodule was categorized into subtypes, including “PFN”, by on-site radiologists. Labels were reviewed centrally, overseen by a single clinician to ensure consistency between sites.
A CNN classifier was trained to produce a score that classifies nodules as (typical) PFN or not, using five-fold cross validation. The PFN classifier was developed by “transfer learning” from an existing benign-vs-malignant AI trained on the US National Lung Screening Trial.
To compare the CNN with human performance, independent validation was performed on a separate dataset of 158 benign patients (196 nodules/nodule images) from two of the sites. Three readers (two radiologists and a radiology resident) were asked to label each nodule as typical PFN, atypical PFN, or non-PFN. To match the AI training procedure, only the typical-PFN labels were used in the reader study, and compared to atypical/non-PFN classified nodules.
Model performance was evaluated by area under the ROC curve (AUC). For the independent validation, Cohen’s kappa was used to measure both the model’s agreement with reader consensus (at least 2 in agreement) and inter-reader agreement. For Cohen’s kappa calculations the CNN score was binarized using a threshold determined from the internal validation data.
Result
The mean cross-validated AUC on the internal dataset was 92% (95% CI = 90.6–92.9). For the independent dataset, the classifier labelled 61/196 (31%) as typical PFNs, and reader consensus gave 45/196 (23%). Versus reader consensus, the AUC of the CNN on the reader-study dataset was 96% (95% CI 93.3–98.4). Both the classifier–reader agreement [(k=0.74) 90%] and the inter-reader agreement [(k=0.64–0.79) 88%-92%] were substantial.
Conclusion
The performance of the PFN classifier is similar to that of radiologists and is within the inter-reader variability of radiologists. This demonstrates the potential utility of CNN-based systems for automatic PFN classification.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.06 - Independent Validation of a Novel High-Resolution Computed Tomography-Based Radiomic Classifier for Indeterminate Lung Nodules (Now Available) (ID 2862)
11:00 - 12:30 | Presenting Author(s): Tobias Peikert | Author(s): Fenghai Duan, Srinivasan Rajagopalan, Ron Karwoski, Aneri B Balar, Dhairya Lakhani, Sanja L Antic, Brian Bartholmai, Johnson Tucker, Pierre P Massion, Fabien Maldonado
- Abstract
- Presentation
Background
Optimization of the clinical management of incidentally- and screen-identified lung nodules is urgently needed to limit the number of unnecessary invasive diagnostic interventions, and therefore morbidity, mortality and healthcare costs. We recently developed and internally validated a novel radiomics-based approach for the classification of screen-detected indeterminate nodules, and present herein validation of this algorithm in an independent cohort.
Method
In a previous study, we developed a multivariate prediction model evaluating independent quantitative variables assessing various radiologic nodule features such as sphericity, flatness, elongation, spiculation, lobulation and curvature. Nodules between 7 and 30 mm of largest diameter were selected from the National Lung Screening Trial (n=726 indeterminate nodules, benign (n = 318) and malignant (n = 408)) were used to derive this model using least absolute shrinkage and selection operator (LASSO) method with bootstrapping method applied for the internal validation. Eight variables capturing vertical location, size, shape, density and surface characteristics were included with an optimism-correct area under the curve (AUC) of 0.94. For this study, an independent dataset of 203 incidentally-identified lung nodules obtained from the indeterminate pulmonary nodule registry at Vanderbilt University was identified. CT datasets were transferred to Mayo Clinic (Rochester, MN) for analysis. Nodules were segmented manually using the ANALYZE software (Biomedical Imaging Resource, Mayo Clinic, Rochester, MN), and radiomic analysis was performed using the 8-variable radiomic diagnostic algorithm derived from the NLST. The Brock model was also used to calculate probability of malignancy for all NLST and Vanderbilt nodules.
Result
Brock scores were calculated for 685 NLST nodules (excluded: interval cancers, n=12; missing values needed for Brock score, n=29). The AUC for the Brock score (AUC Brock) for NLST nodules was 0.83 which was inferior to the AUC for the radiomic model (AUC Radiomic =0.94, P<0.001). When the subset of intermediate pre-test probability of lung cancer was considered (Brock score > 10 but <= 60), the AUC Brock was 0.61 (95% CI: 0.54-0.68) whereas the AUC Radiomic was 0.88 (95% CI: 0.84-0.93). A total of 203 incidentally found pulmonary nodules with available clinical information and biopsy or surgery-proven histology identified in the Vanderbilt indeterminate pulmonary nodule registry, and all histology data and corresponding CT images were reviewed. CT images were transferred to Mayo Clinic for analysis. After exclusion of duplicate CT datasets, unanalyzable CT images and not identifiable nodules (n=27 cases), 176 nodules were segmented and analyzed, including 84 benign and 92 malignant nodules. The AUC was 0.89 (95% CI: 0.85-0.94). For comparison, the AUC Brock was 0.88 (95% CI: 0.83-0.94). When the subset of intermediate pre-test probability of lung cancer was considered (Brock score > 10 but <= 60), the AUC Brock was 0.76 (95% CI: 0.63-0.89) whereas the AUC Radiomic was 0.85 (95% CI: 0.74-0.95).
Conclusion
Our radiomic classifier demonstrates good performance characteristics on an independent retrospective validation dataset. If prospectively validated, integration into clinical decision making algorithm could significantly impact patient care.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.07 - Discrimination of Lung Invasive Adenocarcinoma with Micropapillary Pattern Based on CT Radiomics (Now Available) (ID 399)
11:00 - 12:30 | Presenting Author(s): Haihua Yang | Author(s): Yunyu Xu, Wenbin Ji, Liqiao Hou, Chao Zhou, Wei Wang, Suna Zhou, Feng-Ming Spring Kong
- Abstract
- Presentation
Background
To develop and validate the radiomics nomogram on the discrimination of lung invasive adenocarcinoma (IAC) with micropapillary pattern from non-micropapillary pattern lesion and improve the diagnostic accuracy rate of lung invasive adenocarcinoma with micropapillary pattern before operations and provide guidance for follow-up treatments.
Method
Forty-one pathologically confirmed lung invasive adenocarcinomas with micropapillary pattern from January 2014 to December 2018 were included. Eighty-two pathologically confirmed lung invasive adenocarcinomas without micropapillary pattern from January 2018 to December 2018 were collected. Select 86 patients (70%) randomly from the 123 patients as the primary cohort, and the other 37 patients (30%) were set as an independent validation cohort. Least absolute shrinkage and selection operator (Lasso) was used for feature selection based on contrast enhancement CT images and then radiomics signature building. ROC analysis and AUC were used to value the ability to identify the lung invasive adenocarcinomas with micropapillary pattern.
Result
According to GrayLevelCooccurenceMatrix3, Intensity Histogram and Shape, nine hundred and eighty-five radiomics features were extracted by IBEX. And after data pre-processing such as eliminating missing items, strong correlation variables and multicollinear variables, the features were reduced to 40 features. Based on Mann-Whitney U Test, 28 features were figured out from the 40 features. Then Lasso was used to reduce the features to 3 features (10-1clusterprominenc, -333-4clusterprominence, 8-1contrast) as the most meaningful discriminators to build the radiomics signatures (Table 1). According to SPSS21.0 binary logistic regression analysis, ROC analysis and AUC show that the radiomics signature have effective discrimination performance of lung invasive adenocarcinoma with micropapillary pattern from non- micropapillary pattern lesion (AUC=0.766) and it reflects better in the independent validation cohort (AUC=0.807) (Figure 1).
Table 1 Three characteristic prediction parameters in radiomics label
prediction parameter P value U value W value AUC 10-1clusterprominence <0.005 765.000 4168.000 0.772 -333-4clusterpromise <0.005 790.000 4193.000 0.765 8-1contrast <0.005 919.000 4322.000 0.727
Conclusion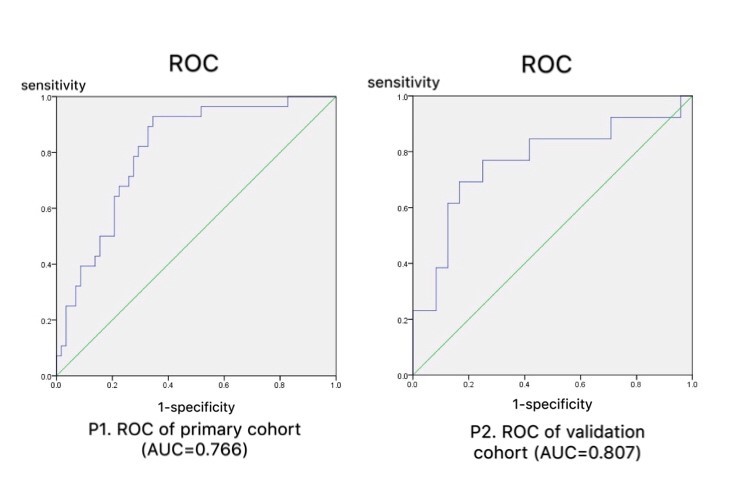
The radiomics signature established in this study have effective prediction of lung invasive adenocarcinoma with micropapillary pattern and non- micropapillary pattern lesion.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
OA06.08 - Discussant - OA06.05, OA06.06, OA06.07 (ID 3757)
11:00 - 12:30 | Presenting Author(s): Jamie Ostroff
- Abstract
Abstract not provided
Author of
-
+
OA10 - Sophisticated TNM Staging System for Lung Cancer (ID 136)
- Event: WCLC 2019
- Type: Oral Session
- Track: Staging
- Presentations: 1
- Now Available
- Moderators:Ke-Neng Chen, Pedro Lopez De Castro
- Coordinates: 9/09/2019, 14:00 - 15:30, Toronto (1985)
-
+
OA10.07 - Concordance Between ROSE and Final Diagnosis in Patients Undergoing EBUS- TBNA for Non-Small Cell Lung Cancer Staging (Now Available) (ID 1123)
14:00 - 15:30 | Author(s): Lluis Esteban Tejero
- Abstract
- Presentation
Background
In patients with non-small cell lung cancer (NSCLC) undergoing mediastinal staging through endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) clinical decision making is based in rapid on-site evaluation (ROSE) findings. We aimed to analyze the accuracy of ROSE determining the rate of concordance between ROSE and final diagnosis.
Method
Prospective study that included patients undergoing EBUS-TBNA for NSCLC staging. Results of ROSE were compared with final diagnosis.
Result
Sixty-four patients were included and 637 lymph nodes (LN) were sampled: a median of 10 (IQR 8-12) LNs and a median of 5 (IQR: 4-6) nodal stations per patient were sampled. The diagnoses of ROSE were concordant with the final diagnoses in 612 (96.1%) cases and non-concordant in 25 (3.9%). There were 7 (1.2%) LNs in which the diagnosis of ROSE was non-malignant with a final diagnosis of malignancy. On the contrary, there was a single case (0.2%) in which the diagnosis of ROSE was malignant with the final diagnosis being normal LN. Considering final diagnosis as the gold standard, the sensitivity, specificity and overall accuracy of ROSE were 98.6, 97.2 and 98.5% respectively.
Table 1:
ConclusionDIAGNOSIS
ROSE
FINAL DIAGNOSIS
Non-diagnostic/
Inadequate
(82)
Benign/
Normal lymph node (520)
Malignancy
(36)
Non-diagnostic/
Inadequate
(70)
67
3
0
Benign/
Normal lymph node (526)
14
511
1
Malignancy
(42)
1
6
35
The preliminary diagnoses (ROSE) are concordant with the final diagnoses in a high percentage. Consequently, clinical decisions based on the ROSE can be taken with confidence.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.


