Virtual Library
Start Your Search
Mark Berry
Author of
-
+
MA08 - Pawing the Way to Improve Outcomes in Stage III NSCLC (ID 127)
- Event: WCLC 2019
- Type: Mini Oral Session
- Track: Treatment of Locoregional Disease - NSCLC
- Presentations: 1
- Now Available
- Moderators:Simon Ekman, Helena A Yu
- Coordinates: 9/08/2019, 15:15 - 16:45, Tokyo (1982)
-
+
MA08.05 - A Multi-Center Analysis of Right vs Left-Sided Pneumonectomy Following Induction Therapy (Now Available) (ID 1178)
15:15 - 16:45 | Author(s): Mark Berry
- Abstract
- Presentation
Background
Previous single-center studies of pneumonectomy following induction therapy for non-small-cell lung cancer (NSCLC) have found a significant perioperative risk associated with right-sided pneumonectomy. We examined the impact of laterality on long-term survival after induction therapy followed by pneumonectomy in a multi-institutional analysis.
Method
Perioperative and long-term outcomes of patients with NSCLC who underwent pneumonectomy following induction chemotherapy with or without radiation from 2000-2016 across 3 institutions were evaluated using multivariable logistic regression, Cox proportional hazards modeling and propensity score-matched analysis. Patients who underwent a completion pneumonectomy or who had M1 disease were excluded from the analysis.
Result
During the study period, 172 patients (right n = 78 [45%], left n = 94 [55%]) met inclusion criteria. Right-sided pneumonectomy was associated with a similar perioperative complication rate (38% [30/78] vs 27% [25/94], p=0.10), and 30-day (13% [10/78] vs 9% [8/94], p=0.36) and 90-day mortality (23% [18/78] vs 13% [12/94], p=0.08) when compared to left-sided pneumonectomy. In multivariable analysis, right-sided pneumonectomy was not found to be a predictor of higher perioperative complications (OR 0.85 [95% CI: 0.33-2.14], p=0.73) or 30-day (OR 2.06 [95% CI: 0.44-9.69], p=0.36) and 90-day mortality (OR 2.06 [95% CI: 0.54-7.88], p=0.29). Overall survival between right and left pneumonectomy was not significantly different in unadjusted (5-year survival 30% [95% CI: 19%-41%] vs 29% [95% CI: 20%-39%], log-rank p=0.77 [Figure]) or multivariable analysis (adjusted hazard ratio, 1.05 [95% CI: 0.63-1.76], p = 0.84). A propensity score-matched analysis of 108 patients balancing baseline characteristics—including pulmonary function, tumor size and stage—was also performed, and found no significant differences in perioperative complication rates (46% [25/54] vs 48% [26/54], p=0.85), 30-day (17% [9/54] vs 7% [4/54], p=0.14) and 90-day mortality (26% [14/54] vs 13% [7/54], p=0.09) between right versus left pneumonectomy, respectively. Overall survival was not significantly different between right- and left-sided pneumonectomy (5-year survival 33% [95% CI: 20%-47%] vs 28% [95% CI: 16%-41%], log-rank p=0.98).
Conclusion
In this multi-center analysis, right-sided pneumonectomy after induction therapy was not associated with significantly higher perioperative mortality rates or worse long-term survival when compared to a left-sided pneumonectomy.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
P2.05 - Interventional Diagnostic/Pulmonology (ID 168)
- Event: WCLC 2019
- Type: Poster Viewing in the Exhibit Hall
- Track: Interventional Diagnostics/Pulmonology
- Presentations: 1
- Moderators:
- Coordinates: 9/09/2019, 10:15 - 18:15, Exhibit Hall
-
+
P2.05-01 - Broad Genomic Profiling of Bronchoalveolar Lavage Fluid in Lung Cancer (ID 615)
10:15 - 18:15 | Author(s): Mark Berry
- Abstract
Background
We hypothesized that tumor-derived mutations from non-small cell lung cancer (NSCLC) are readily detectable in bronchoalveolar lavage (BAL). To explore our hypothesis, we employed the CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) method to identify somatic mutations in BAL compared to blood.
Method
We profiled 200 matching lavage, plasma, and PBMC samples from a total of 38 NSCLC patients and 21 controls. We first applied a tumor-informed calling approach to most sensitively detect mutations in BAL and plasma. We then applied a tumor-naïve mutation calling strategy to explore the effect of field cancerization in at risk patients with lung nodules or who smoked. Last, we developed a BAL mutation classifier to differentiate patients with cancer from those without and compared the performance of this classifier to BAL cytology.
Result
Tumors were primarily lung adenocarcinomas (84%) and mostly early stage disease (I-II 71%; III-IV 29%). We called a median of 4 mutations per tumor. TP53 and KRAS were the most frequently detected variants in tumor (47% and 35% respectively) and lavage cell free (cf) DNA (38% and 26% respectively). Using a tumor-informed approach, we detected significantly more variants in lavage cfDNA than in plasma from cancer patients (p<0.001) and variants were more frequently called in lavage cfDNA than in plasma from cancer patients (77% vs. 41%, p=0.004). As expected, tumor-naïve calling resulted in fewer variants detected in both sample types when compared to tumor-informed calling but we identified more tumor mutations (p<0.001) and more putative cancer driver mutations (p< 0.003) in lavage cfDNA than in plasma. Mutations of cancer driver genes at the patient level and average %VAF at the gene level were significantly lower in lavage cfDNA controls compared to cancer patients (p=0.017 and p=0.016 respectively). Since we also detected mutations in controls, presumably secondary to field cancerization and somatic mosaicism, we developed a risk score of mutation features to classify whether a BAL specimen was likely to come from a cancer patient or control. At a risk score level that identified all non-cancers as benign (A), this BAL classifier of 11 mutation features identified more cancers than cytology for all stages of lung cancer (65% vs 12%, p=0.001) and in stage I/II disease (50% vs. 20%, p=0.25; B).
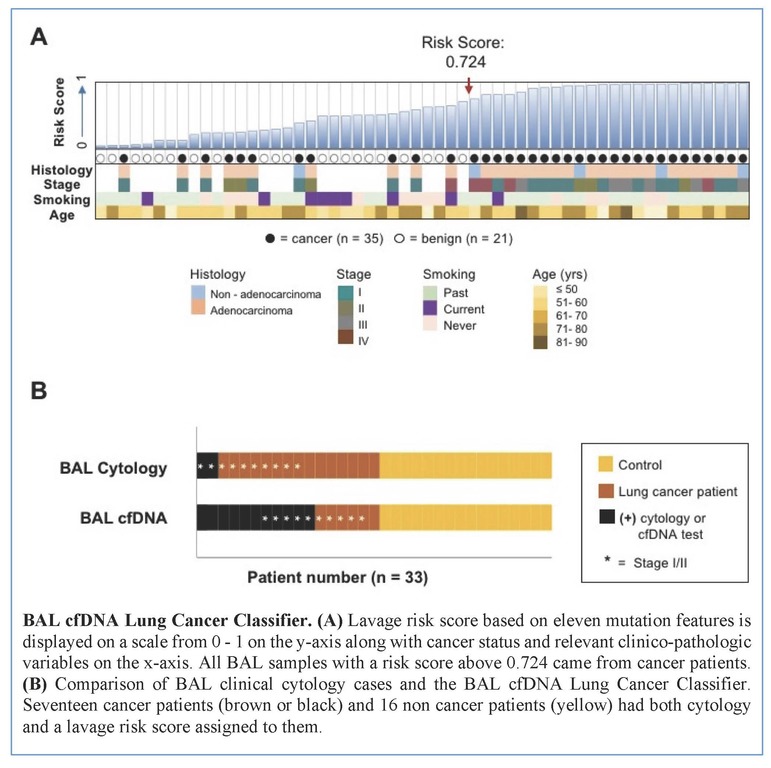
Conclusion
We show here that BAL genomic profiling may augment plasma profiling and BAL cytology for diagnosing and profiling NSCLC. Validation studies will be required to confirm our findings.


