Virtual Library
Start Your Search
Michael PA Davies
Author of
-
+
FP07 - Pathology (ID 109)
- Event: WCLC 2020
- Type: Posters (Featured)
- Track: Pathology, Molecular Pathology and Diagnostic Biomarkers
- Presentations: 1
- Moderators:
- Coordinates: 1/28/2021, 00:00 - 00:00, ePoster Hall
-
+
FP07.02 - Deep Learning Based Analysis of Multiplex IHC Accurately Interprets PD-L1 and Provides Prognostic Information in NSCLC (ID 1044)
00:00 - 00:00 | Author(s): Michael PA Davies
- Abstract
Introduction
Most assessment of PD-L1 expression by immunochemistry to guide immuno-modulatory (IM) therapy in non-small cell lung cancer (NSCLC) requires only tumour cells to be scored, the so-called ‘tumour proportion score’ (TPS). This narrow approach ignores what is almost certainly crucial information within the tumour microenvironment (TME), particularly the nature and distribution of tumour infiltrating lymphocytes (TILs). Such features of the TME are underutilised in the clinical setting and are strong candidates for improving the predictive power of PD-L1 assessment alone. We describe a novel approach to analysing the TME in this context.
Methods
Consecutive whole slide sections from 92 resected NSCLCs were stained with H&E and immunolabelled for PD-L1 alone using the SP263 clone (monoplex), PD-L1 using the SP142 clone, CD68 and CD3 (triplex), and FoxP3, PD-1 and CD8 (triplex). Monoplex PD-L1 expression was scored by two pathologists to generate a consensus TPS score and categorised as negative, weak or strong expression; <1%, 1-49% and ≥50% TPS respectively. An existing deep-learning based PD-L1 solution was used to automatically score PD-L1 TPS. For the triplex images, densities of ‘positive’ cells were computed automatically using assay-specific deep learning algorithms, with a separate deep learning algorithm used to segment epithelial regions. The density of PD-L1+ve/CD3-ve/CD68-ve cells was used as a surrogate PD-L1 TPS with 33rd and 66th quartiles defining clinical group categorisation. Survival data were used in Kaplan-Meier survival analysis of groups divided by PD-L1 expression and immune cell densities.
Results
TPS scores as a continuous variable correlated well between pathologist assessment and both monoplex (Pearson correlation coefficient 0.977) and triplex (Pearson CC 0.849) assessments. Automated interpretation via triplex was similar to monoplex for grouping samples by dichotomous division at a 50% cut-off (91.2% vs. 94.6% of cases) and for placement into clinically relevant categories (79.1%, Cohen’s kappa coefficient Κ = 0.687 vs. 85.9%, K = 0.786). Sub-group analysis of tumours divided by the median for each variable into ‘high’ or ‘low’ revealed no significant difference in overall survival (OS) when stratified by CD3, FoxP3, PD-1 or CD68. However, high CD8+ve TIL densities and strong PD-L1 expression both correlated with improved OS (56 vs. 39 months, p=0.028; 60 vs. 41 months, p=0.035 respectively). In addition, tumours with a PD-L1 high/CD8+ve high profile showed significantly better OS than those assessed as PD-L1 low/CD8+ve (57 vs. 36 months, p=0.019).
Conclusion
Automated, deep-learning based, algorithmic scoring of PD-L1 expression is a valid and accurate approach to its assessment, and utilising triplex data provides important prognostic information. Discrepancies between monoplex and triplex assessment might be attributed to the different anti-PD-L1 antibody clones used, but the automated nature of triplex that excludes macrophages and TILs still performs very well. Our study shows the power of using this approach to augment the power of PD-L1 expression alone as a predictor of response to IM therapy and to provide prognostic information.
-
+
FP09 - Screening and Early Detection (ID 175)
- Event: WCLC 2020
- Type: Posters (Featured)
- Track: Screening and Early Detection
- Presentations: 1
- Moderators:
- Coordinates: 1/28/2021, 00:00 - 00:00, ePoster Hall
-
+
FP09.02 - Calibration of the LLP Lung Cancer Risk Stratification Model: Prospective Validation in the UKLS Cohort of 76,000 People (ID 3137)
00:00 - 00:00 | Author(s): Michael PA Davies
- Abstract
- Presentation
Introduction
Introduction: Evidence from two large randomised trials estimates a significant 20% -24% reduction in lung cancer mortality in the low dose CT screening arm.
Future lung cancer screening programmes require identification of high-risk populations to optimise detection in those who would benefit from screening for lung cancer and provide a cost-effective programme. The Liverpool Lung Project risk model (LLPv1) was first developed in 2008, validated in three international datasets in 2012 and amended in the UKLS trial (LLPv2). We report on further amendments to the model (LLPv3), where it is calibrated to the whole of England and more recent lung cancer incidence data.
Methods
75,958 participants responded to the first approach UKLS trial questionnaire. We obtained cancer registration data from Public Health England, linked through a third party, providing at least 5 years of follow-up data. A ROC curve was plotted to assess ability of the LLP model to distinguish between those individuals who went on to develop lung cancer. In order to assess the calibration of the predictive model, we ranked the LLPv3 risk scores, assigned decile groups, and carried out a linear regression of the log-odds () of the rate of observed cancers in each group against the log-odds of the mean of the risk scores in the group (See Figure1).
Results
The ROC curves for LLPv2 and LLPv3 are almost identical, as would be expected, since the coefficients pertaining to the risk factors are the same; ranking of most individuals is unchanged from LLPv2 to LLPv3, thus, the discrimination is very similar. The AUC in each case was 0.81 (95% CI 0.79 - 0.82). After excluding individuals allocated to the UKLS trial intervention arm, the AUC and 95% confidence interval were also unchanged.
The LLPv3 model also provided a very good estimation of future risk through comparison of observed and expected lung cancer outcomes. However, without the calibration to the current England population in terms of age and sex specific lung cancer risk, there would have been considerable overestimation of risk. This is consistent with the findings of Katki et al. [Ann Intern Med 2018;169(1):10-19], who concluded that previous versions of the LLP model overestimated future incidence.
Conclusion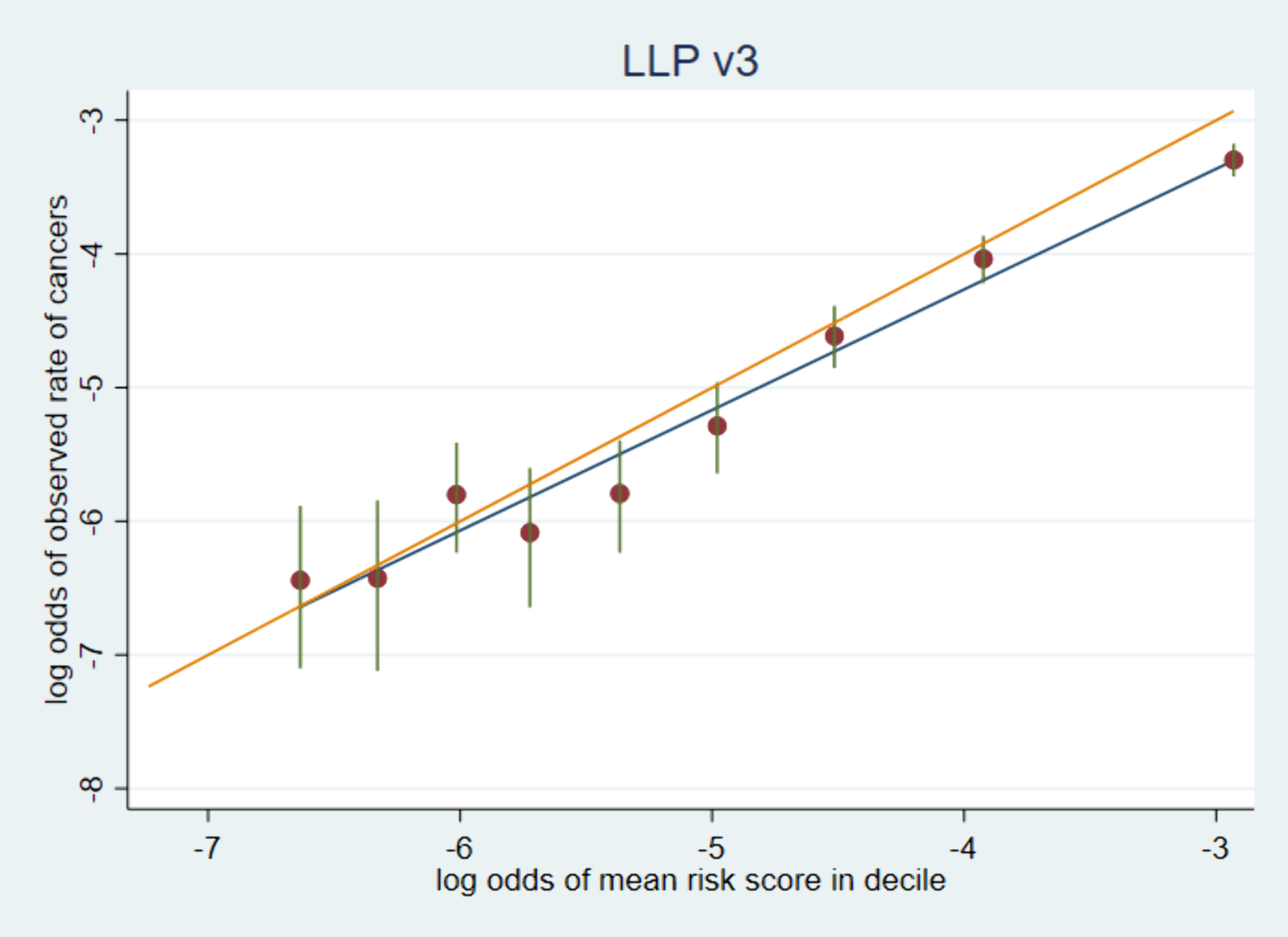
The discrimination of LLPv2 and LLPv3 was excellent. LLPv3, which was calibrated to contemporary, English incidence, achieved more accurate prediction of absolute incidence, and would be more effective in selecting a high-risk group for surveillance in England today.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.


