Virtual Library
Start Your Search
Max Diehn
Moderator of
-
+
ES25 - Liquid Biopsy (ID 27)
- Event: WCLC 2019
- Type: Educational Session
- Track: Treatment of Early Stage/Localized Disease
- Presentations: 4
- Now Available
- Moderators:Max Diehn, Lucia Milla Collado
- Coordinates: 9/10/2019, 14:30 - 16:00, Colorado Springs (1994)
-
+
ES25.01 - Liquid Biopsy: State of the Science (Now Available) (ID 3287)
14:30 - 16:00 | Presenting Author(s): Eloisa Jantus-Lewintre
- Abstract
- Presentation
Abstract
I. The present.
Molecular diagnosis in cancer certainly requires the analysis of a tumor biopsy. However, in lung cancer, there is still a 20- 30% of tissue failure rates for tumor genotyping in routine pathological samples. As a consequence, liquid biopsy (LB) has emerged as a valid alternative source of information for the analysis of tumor specific alterations. LB refers to specimens obtained from body fluid such as blood, urine, saliva, cerebrospinal fluid, among others.
In the complex matrix represented by blood, the main clinical developments have focused on the analysis of: i) circulating tumor DNA (ctDNA), which represents a small part of cell free circulating DNA released from tumor cells and, ii) circulating tumor cells (CTCs), defined as disseminated cancer cells in the bloodstream. Each of these materials offers unique opportunities to test different biomarkers and to analyze characteristics of the tumors.
The advantages of the use of blood samples are clear: i) it is a minimally invasive way to get relevant tumor information, ii) serial samples can be obtained capturing tumor evolution in real time, iii) LB abrogates the limitations associated with tumor heterogeneity, since nucleic acids or tumor cells present in circulation recapitulate the information belonging from different tumor locations (primary tumor and metastases), iv) the development of new sensitive assays for analyses of ctDNA and CTCs allow the assessment of minimal residual disease and v) the costs of LB analysis are comparable with other molecular biology techniques already used in the clinical setting in addition to the reduced risks of complications associated with tissue biopsy. All these factors accelerated the implantation of LB in the clinical practice in oncology in several scenarios, especially in lung cancer.
I.1.Lung Cancer. Clinical applications of liquid biopsies.
At present, LB is no longer a promise but a reality allowing better treatment selection, real-time monitoring of lung cancer patients and early detection of acquired resistances. Figure 1 highlights the biological basis of LB as a source for biomarkers analysis and key clinical applications in lung cancer.
•Personalized therapeutics/ Resistance detection: regarding the detection of tumor-associated genetic alterations in LB samples, there are a lot of scientific data demonstrating similar response rates to targeted therapies than the obtained in tissue biopsies. In particular, in the context of NSCLC patients with progressive or recurrent disease during treatment with TKIs, the IASLC guidelines suggest the use of LB-first algorithm to detect resistance mechanism.
For those lung cancer patients receiving immune-based therapeutic treatments, recent data show that assessment of tumor mutational burden in plasma (bTMB) correlated with the values found in tissue and predicts efficacy of immune-checkpoint inhibitors.
•Detection of minimal residual disease: Persistent detection of ctDNA or CTCs after local therapy or after adjuvant treatments was found associated with poor clinical outcome. In this particular clinical setting it is important to highlight that sensitivity of the methods used for ctDNA or CTCs evaluation really matters.
•Real-time monitoring of disease: this is one of the most interesting application of LB, since tissue biopsies are intrinsically unable to capture tumor heterogeneity while ctDNA can comprehensively recapitulates clonal evolution over time, allowing to early detect and track the emergency of resistance mutations.
II. The future.
Current assays for LB analysis do not meet all the needs required for the fully implementation of the Precision Oncology. There is still room for improvement to reach its maximum informative potential. Hopefully, studies on exosomes, platelets, cfRNAs, metabolites, will help to have a more integrative picture of tumor status at each time it is evaluated.
One of the clinical applications in which LB is called to play a key role is in the screening and early detection of lung cancer. In this regard, there are some interesting data coming from multiparametric (DNA and protein) plasma analysis. However, caution is required since there are still some important issues, such as clonal hematopoiesis, that need to be further considered.
Another important challenge for LB is standardization. It is necessary to cross-validate platforms, standardize pre-analytical issues, compare sensitivity of different methodological approaches and also to work in the harmonization of bioinformatic tools for data analysis.
It is clear that in the near future, tests based on the analysis of "liquid biopsies" will be more generalized, offering complementary information to tissue biopsies and providing valuable information to early diagnose lung cancer, to detect molecular progressions even prior to radiographic or clinical progression and as a source for real-time treatment monitoring.
References
Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6(5): 479-91.
Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell. 2017; 31(2):172-179.
Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24
Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016; 5(5):466-482
Heitzer E, Haque IS, Roberts CE, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71-88.
Pantel K, Alix-Panabières C. Liquid biopsy: Potential and challenges. Mol Oncol. 2016; 10(3):371-3.
Rossi G, Ignatiadis M. Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res. 2019;79(11):2798-2804.
Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531-.548

Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
ES25.02 - Liquid Biopsy: Utility for Surveillance in Early Stage (Now Available) (ID 3288)
14:30 - 16:00 | Presenting Author(s): Lecia Sequist
- Abstract
- Presentation
Abstract not provided
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
ES25.03 - Leveraging the Quantitative Nature of Cfdna Genotyping for Lung Cancer Care (Now Available) (ID 3289)
14:30 - 16:00 | Presenting Author(s): Cloud P Paweletz
- Abstract
- Presentation
Abstract
Nearly 70 years after the discovery of free-floating DNA in the blood, also known as cell free DNA (cfDNA) (1), plasma genotyping of cfDNA is transforming cancer care with promise in non-invasive genotyping, early diagnosis, and disease prognostication by detection of minimal residual disease (MRD). At present the most widespread use of cfDNA in lung cancer is detection of mutations in EGFR and KRAS or re-arrangements in ALKand ROS1 in the metastatic setting. In fact, the only FDA approved liquid biopsy test among all cancers is Roche’s Cobas plasma EGFR mutation test for non-small cell lung cancer. (NSCLC) (2), while many diagnostic companies have developed PCR or next generation sequencing (NGS) laboratory developed tests that are commonly reimbursed by payors in house. Next-generation sequencing (NGS) permits broader inquiries, allowing assessment of the mutation status of thousands to millions of bases.
The use of cfDNA in early cancers is confounded by the fact that early detection demands ultra-sensitive assays of low abundant biological markers. As proof of concept Bettegowda et al. performed digital PCR on cfDNA of 640 cancer patients of varying cancer type and stage. Intriguingly they found that rates of tumor DNA differed by tissue of origin, and 47% of stage I cancers and 55% of stage II cancers had detectable circulating tumor DNA (ctDNA) (3). An observation that as we and others have confirmed is a main source for false negatives (4). At present, NGS efforts are focused either on targeted approaches using either barcoded targeted amplicon (TAmSeq) or hybrid capture approaches (CAPP-Seq) covering 10Mb to 50Mb at reported sensitivities of 0.01% to 0.50% for fit for purpose build NGS assays. However, pushing assay sensitivity increases false positives.
Recently, we and other have found that false positives are reported in many commercial assays and are routinely attributed to ‘tumor heterogeneity’ . These can be attributed to DNA shed from normal cells, including germline variants or non-cancerous somatic variants from clonal hematopoiesis (CH) (5-8). The latter is particularly challenging because CH can involve cancer-associated genes (e.g. TP53, JAK2, KRAS). To limit false positives and to investigate the common link between cancer-related mutations within the blood and underlying malignancies broad sequencing of cfDNA should also detect other cancer-related mutations, such as inactivating mutations in tumor suppressors and include assaying of the patients blood cells to filterout germlines. Thus, deep and broad sequencing could provide sensitivity needed to detect low levels of cfDNA alterations in early stage patients. Indeed, Abbosh et al. perform multi region whole-exome sequencing of early-stage NSCLC tumors to show an abundance of clonal mutations in these tumors (9). Abbosh and colleagues provide an intriguing solution to this issue by requiring the detection of two or more SNVs for the determination of the presence of cancer.In this presentation we present factors affecting ultras sensitive assays with particular emphasis on interpretation of commercial tests and future use of cfDNA assays in early cancers.
1. Mandel P and Metais P. Les acides nucleiques du plasma sanguin chez l’homme [in French]. C R Seances Soc Biol Fil 1948;142:241-243.
2.Center for Drug Evaluation and Research. Approved Drugs - cobas EGFR Mutation Test v2 [Internet]. U S Food and Drug Administration Home Page. Center for Drug Evaluation and Research; Available from: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm
3.Bettegowda C, Sausen M, Leary R, Kinde I, Agrawal N, Bartlett B, et al. Detection of Circulating Tumor DNA in Early and Late Stage Human Malignancies. Science Translational Medicine 2014;16:224ra24.
4. Oxnard GR, Thress KS, Alden RS, et al: Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. Journal of Clinical Oncology 34:3375–3382, 2016
5.Hu Y, Ulrich BC, Supplee J, et al: False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res 24:4437-4443, 2018
6.Hu Y, Alden RS, Odegaard JI, et al: Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients. Clin Cancer Res 23:7351-7359, 2017
7.Slavin TP, Banks KC, Chudova D, et al: Identification of Incidental Germline Mutations in Patients with Advanced Solid Tumors Who Underwent Cell-Free Circulating Tumor DNA Sequencing. Journal of Clinical Oncology 36:3459-3465, 2018
8.Oxnard GR, Tara M, Earl H, et al. Genome-wide sequencing for early stage lung cancer detection from plasma cell-free DNA (cfDNA): The Circulating Cancer Genome Atlas (CCGA) study. Journal of Clinical Oncology : LBA8501-LBA8501, 2018
9.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA Analysis Depicts Early Stage Lung Cancer Evolution. Nature 2017;545:446-451.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
ES25.04 - Liquid Biopsy: Utility for Early Detection (Now Available) (ID 3290)
14:30 - 16:00 | Presenting Author(s): Ana Vivancos
- Abstract
- Presentation
Abstract
Personalized treatment according to molecular profile is standard of care in advanced NSCLC patients. Epidermal Growth Factor Receptor (EGFR) activating mutations predict sensitivity to first- and second-generation anti-EGFR tyrosine kinase inhibitors (TKIs) in patients with non-small cell lung cancer (NSCLC). However, obtaining a tissue biopsy remains a limitation in NSCLC patients. Liquid biopsy is a non-invasive method that allows the detection and quantification of tumor somatic mutations in plasma, although around 20% of all tumors don’t appear to shed DNA into the bloodstream. Here, we aimed to correlate the presence and amounts of ctDNA in plasma of NSCLC patients with several clinical parameters, as well as obtaining serial data on non-shedding patients throughout their course of illness.
We collected 280 serial blood samples of 40 patients with NSCLC diagnosis harboring EGFR sensitizing mutations in their tumor biopsy. Extracted ctDNA was tested for five common EGFR mutations (exon 19 deletion, L858R, L861Q, T790M and C797S) by highly sensitive and quantitative Droplet Digital PCR (ddPCRTM; Bio-Rad), at a limit of detection between 0.1-0.5% and quantified the percentage of mutant alleles of EGFR. Of these patients, 16 provided one plasma sample (32%) and the other 34 (68%) provided multiple blood collections with an average of 5 follow-up plasma samples. All patients received targeted TKI therapy before or during the study. Lines of treatment, Progression free survival (PFS) and overall survival (OS) were annotated for each patient in the cohort.
Out of the 40 patients evaluated, we detected presence of baseline ctDNA in 32 patients (80%). Such parameter was independent of the sensitizing mutation; tumors harboring exon 19 or exon 21 mutation tend to equally shed DNA into the bloodstream (78% and 80%, respectively). After a median follow up of 36.1 months, and immature survival data, the PFS and OS are higher among patients with baseline ctDNA positive compared to patients without ctDNA detected: 22 months vs. 13.6 months and 35 vs. 24 months, respectively. T790M and C797S resistance mutations were detected at different prevalences, depending on the TKI treatment regimen, and were always subclonal in plasma as compared to the clonal EGFR mutation (indel 19/L858R). We observed that, for the whole period analyzed in non-shedding patients, ctDNA was never detected.
In our series, sensitizing EGFR mutations in plasma were identified in 80% of the patients by ddPCR. Acquired resistance mutations in EGFR appeared to be subclonal, which might impact detection in liquid biopsy. Shedding is a complex biological entity that warrants further research in order to improve our understanding on its impact in prognosis.
Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
Author of
-
+
P2.05 - Interventional Diagnostic/Pulmonology (ID 168)
- Event: WCLC 2019
- Type: Poster Viewing in the Exhibit Hall
- Track: Interventional Diagnostics/Pulmonology
- Presentations: 1
- Moderators:
- Coordinates: 9/09/2019, 10:15 - 18:15, Exhibit Hall
-
+
P2.05-01 - Broad Genomic Profiling of Bronchoalveolar Lavage Fluid in Lung Cancer (ID 615)
10:15 - 18:15 | Author(s): Max Diehn
- Abstract
Background
We hypothesized that tumor-derived mutations from non-small cell lung cancer (NSCLC) are readily detectable in bronchoalveolar lavage (BAL). To explore our hypothesis, we employed the CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) method to identify somatic mutations in BAL compared to blood.
Method
We profiled 200 matching lavage, plasma, and PBMC samples from a total of 38 NSCLC patients and 21 controls. We first applied a tumor-informed calling approach to most sensitively detect mutations in BAL and plasma. We then applied a tumor-naïve mutation calling strategy to explore the effect of field cancerization in at risk patients with lung nodules or who smoked. Last, we developed a BAL mutation classifier to differentiate patients with cancer from those without and compared the performance of this classifier to BAL cytology.
Result
Tumors were primarily lung adenocarcinomas (84%) and mostly early stage disease (I-II 71%; III-IV 29%). We called a median of 4 mutations per tumor. TP53 and KRAS were the most frequently detected variants in tumor (47% and 35% respectively) and lavage cell free (cf) DNA (38% and 26% respectively). Using a tumor-informed approach, we detected significantly more variants in lavage cfDNA than in plasma from cancer patients (p<0.001) and variants were more frequently called in lavage cfDNA than in plasma from cancer patients (77% vs. 41%, p=0.004). As expected, tumor-naïve calling resulted in fewer variants detected in both sample types when compared to tumor-informed calling but we identified more tumor mutations (p<0.001) and more putative cancer driver mutations (p< 0.003) in lavage cfDNA than in plasma. Mutations of cancer driver genes at the patient level and average %VAF at the gene level were significantly lower in lavage cfDNA controls compared to cancer patients (p=0.017 and p=0.016 respectively). Since we also detected mutations in controls, presumably secondary to field cancerization and somatic mosaicism, we developed a risk score of mutation features to classify whether a BAL specimen was likely to come from a cancer patient or control. At a risk score level that identified all non-cancers as benign (A), this BAL classifier of 11 mutation features identified more cancers than cytology for all stages of lung cancer (65% vs 12%, p=0.001) and in stage I/II disease (50% vs. 20%, p=0.25; B).
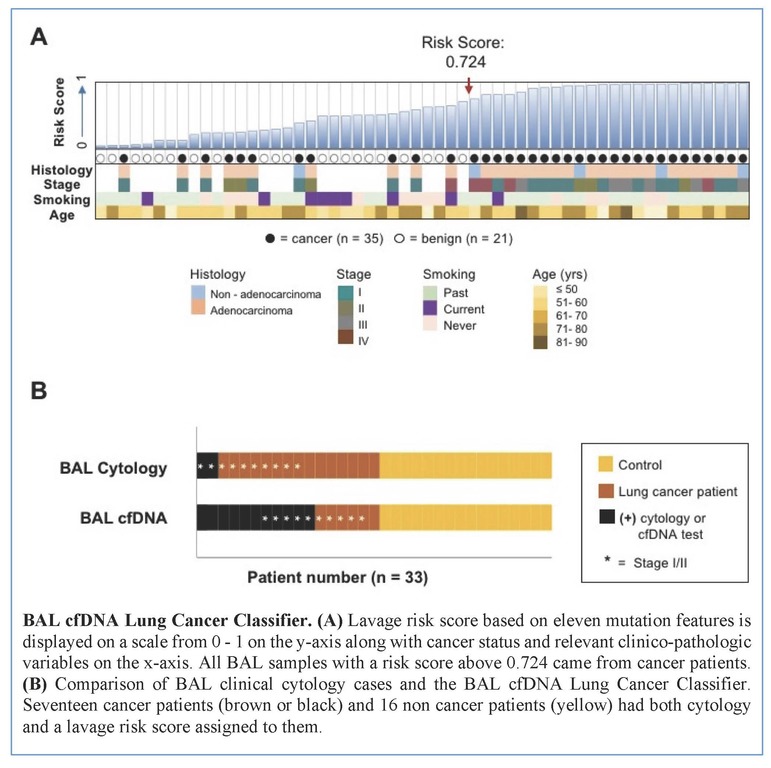
Conclusion
We show here that BAL genomic profiling may augment plasma profiling and BAL cytology for diagnosing and profiling NSCLC. Validation studies will be required to confirm our findings.


