Virtual Library
Start Your Search
Jian Su
Author of
-
+
JCSE01 - Perspectives for Lung Cancer Early Detection (ID 779)
- Event: WCLC 2018
- Type: Joint IASLC/CSCO/CAALC Session
- Track: Screening and Early Detection
- Presentations: 1
- Moderators:
- Coordinates: 9/23/2018, 07:30 - 11:15, Room 202 BD
-
+
JCSE01.22 - Differential Molecular Mechanisms Associated with Dramatic and Gradual Progression in NSCLC Patients with Intrathoracic Dissemination (ID 14713)
11:15 - 11:15 | Author(s): Jian Su
- Abstract
Background
Lung cancer is a highly heterogeneous disease with diverse clinical outcomes. The pleural cavity is a frequent metastasis site of proximal lung cancer. Better understanding of its underlining molecular mechanisms associated with dramatic and gradual progression of pleural metastasis in patients with non-small cell lung cancer (NSCLC) is essential for prognosis, intervention and new therapy development.We performed whole-exome sequencing (WES) of matched primary lung adenocarcinoma and pleural metastatic tumors from 26 lung cancer patients with dramatic progression (DP, n=13) or gradual progression (GP, n=13). Somatic alterations at both genome-wide level and gene level were detected. Kaplan-Meier survival analysis and multivariate Cox regression models were applied to analyze the association between different somatic alterations and clinical parameters.We first analyzed the differences in somatic alterations between AP and RP group in the primary tumors, and identified higher somatic copy number alteration (SCNA) level in DP group compared to GP group, which is significantly (p=0.016) associated with poorer progression-free survival (PFS). More specifically, patients with chromosome 18q loss in the primary tumor showed a trend (p=0.107) towards poorer PFS. PTEN (p=0.002) and GNAS (p=0.002) mutations are enriched in the primary tumors of DP group, and are associated with poorer PFS. Furthermore, pleural metastatic tumors harbor a relatively higher level of mutation burden (p=0.105) and significantly increased SCNA (p=0.035) compared to the primary tumors.NSCLC patients in the attenuated progression group have more stable genomes. High level of genomic instability, GNAS and PTENmutations, as well as chromosome 18q loss are associated with rapid progression. a9ded1e5ce5d75814730bb4caaf49419
-
+
MA24 - Genomic Evolution, KEAP 3 and More Non-Coding RNA (ID 928)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Biology
- Presentations: 1
- Moderators:
- Coordinates: 9/26/2018, 10:30 - 12:00, Room 205 BD
-
+
MA24.01 - Genomic Evolution Trajectory Depicts Invasiveness Acquisition from Pre-invasive to Invasive Adenocarcinoma (ID 11840)
10:30 - 10:35 | Author(s): Jian Su
- Abstract
- Presentation
Background
Accumulation of molecular abnormalities may depict evolution trajectories of tumor initiation and development. However, the genomic profile of early stage adenocarcinoma and molecular mechanism of invasiveness acquisition from pre-invasive to invasive adenocarcinoma remains barely explored.
a9ded1e5ce5d75814730bb4caaf49419 Method
We simultaneously collected 20 patients with adenocarcinoma in situ (AIS) (n=5), minimally invasive adenocarcinoma (MIA) (n=5) and stage IA adenocarcinoma (lepidic/acinar predominant) (n=10). Whole exon sequencing (WES) was performed in pre-invasive adenocarcinoma with multi-region specimens and stage IA adenocarcinoma. Analysis of genomic alteration among different pathological status was performed and tumor mutation burden (TMB) was calculated as well as six mutation types individually. Enriched pathways of each pathology were measured through KEGG analysis.
4c3880bb027f159e801041b1021e88e8 Result
Baseline characteristics was generated through heatmap with smokers (2/20, 10%) and EGFR mutation (13/20, 65%) among whole population. AIS/MIA indicated much lower number of mutations than invasive adenocarcinoma (IAC) while TMB revealed the same trend without statistical significance. Multi-region sequencing showed high heterogeneity of single nucleotide variation (SNV) in AIS and MIA. Unique SNV presented dominant proportion in initial status. Cluster analysis showed higher copy number variation in AIS/MIA than IAC with cell adhesion molecules (CAMs) enriched in AIS/MIA while variety pathway enrichment in IAC through KEGG analysis. C>A transversions held major proportion in early stage adenocarcinoma and a significant increase in the proportion of C>T and C>G mutation was exhibited when evolving into IAC.
8eea62084ca7e541d918e823422bd82e Conclusion
Intratumor heterogeneity may occur in the very beginning of adenocarcinoma. High copy number variation was dominant event for AIS/MIA while higher tumor mutation burden was seen in IAC. Tobacco signature encompassing C>A transversions dominates the early development of adenocarcinoma and APOBEC signature may play a potential role in acquisition of cancer invasiveness.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
P1.11 - Screening and Early Detection (Not CME Accredited Session) (ID 943)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.11-18 - A Classification-Based Machine Learning Method Reveals Exosomal miRNA Biomarkers for Patients with Pulmonary Ground Glass Nodule (ID 12462)
16:45 - 18:00 | Author(s): Jian Su
- Abstract
Background
Non-invasive detection of lung cancer is of critical importance but has proven challenging due to the rate of false-negative diagnosis with current tests. Plasma exosomes have been implicated as a non-invasive diagnostic source. However, little high throughput screening has been done in the early-stage lung cancer and problems such as bias of enrollment, less rigorous identification exists. This study aimed to reveal the plasma exosome-derived miRNA biomarkers for early-stage lung cancer patients, especially those with ground glass nodule (GGN).
a9ded1e5ce5d75814730bb4caaf49419 Method
Pre-operative and paired post-operative plasma samples from patients with solitary pulmonary nodule and healthy volunteers were prospectively collected. Finally 38 malignant nodules, 7 benign nodules and 5 healthy volunteers were enrolled. The malignant nodules included 9 pure GGNs, 11 mixed GGNs and 18 solid nodules. Exosomes were collected from 1mL plasma and were isolated with 3D Medicine EV isolation kit. Exosomal miRNA profiling was performed using miRNA-seq. And an exosomal miRNA diagnostic model for patients with malignant nodules was constructed by using support vector machine (SVM).
4c3880bb027f159e801041b1021e88e8 Result
In general, malignant nodules, benign nodules and healthy volunteers were indistinguishable based on overall clustering. Regarding to malignant nodules, pure GGNs and solid nodules could be separated under principal component analysis (PCA), and the mixed GGNs presented a transitional state between the pure GGNs and the solid nodules. Ultimately, a two-dimensional SVM diagnostic model for discriminating malignant and benign nodules was established. The optimal miRNA combination could reach an area under curve (AUC) of 0.96, with sensitivity and specificity of 94.7% and 91.7%, respectively.
8eea62084ca7e541d918e823422bd82e Conclusion
This preliminary analysis highlights the potential of exosomal miRNA based liquid biopsy for non-invasive detection of early-stage lung cancer. The SVM model seems could effectively distinguish pulmonary nodules, but needs further verified.
6f8b794f3246b0c1e1780bb4d4d5dc53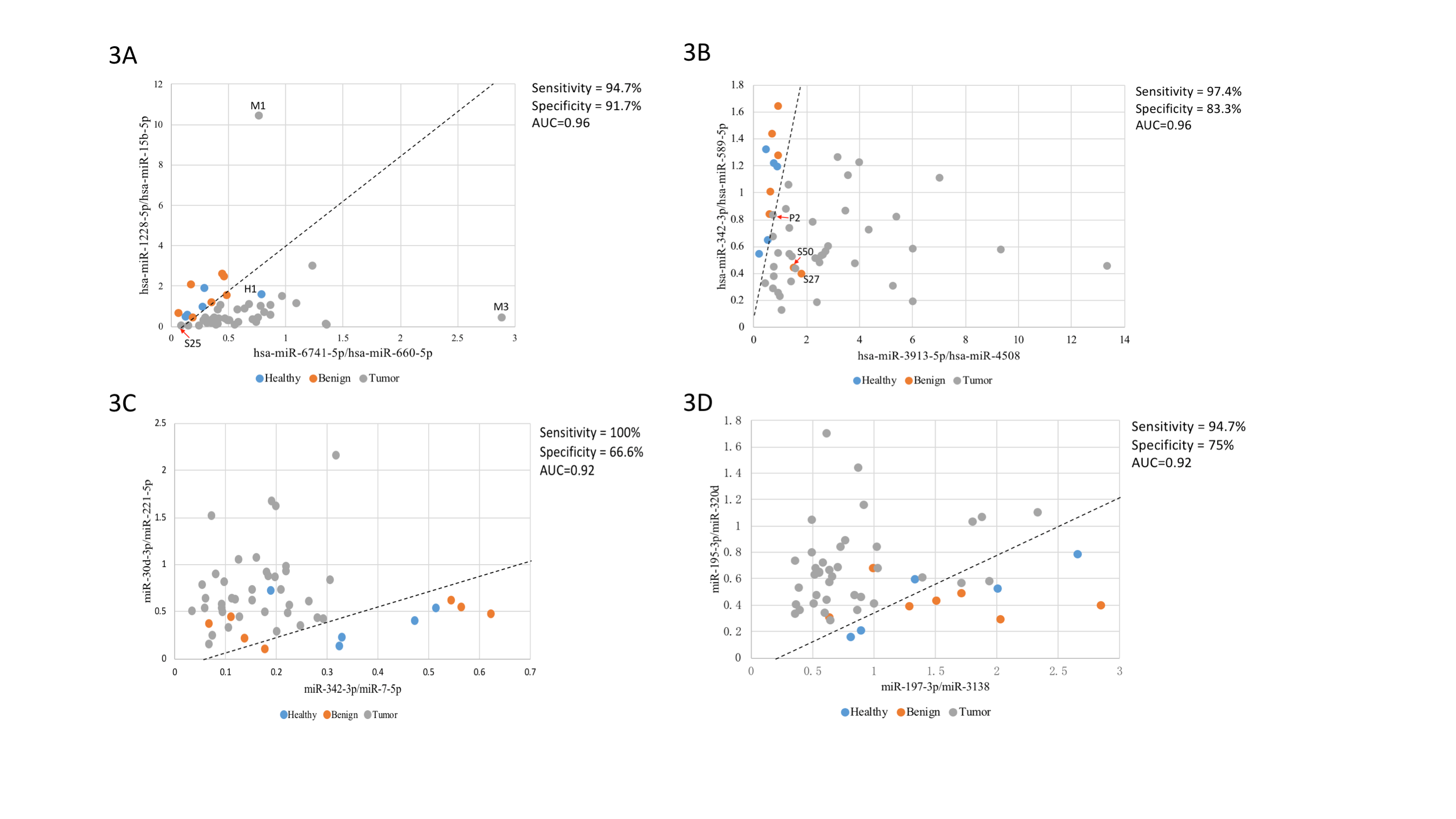
-
+
P3.12 - Small Cell Lung Cancer/NET (Not CME Accredited Session) (ID 978)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/26/2018, 12:00 - 13:30, Exhibit Hall
-
+
P3.12-10 - Immunogenomic Characteristics of SCLC and LCNEC Redefined Molecular Subgroups (ID 12577)
12:00 - 13:30 | Author(s): Jian Su
- Abstract
Background
While small-cell lung cancer (SCLC) and large-cell neuroendocrine carcinoma (LCNEC) are distinct classes of high-grade neuroendocrine carcinomas, the differential diagnosis between SCLC and LCNEC remains challenging. In fact SCLC and LCNEC overlap in clinical, histopathologic, cytologic, morphologic and genetic characteristics. Molecular profiling with microarray or next-generation sequencing has provided growing evidence suggesting that both SCLC and LCNEC are biologically heterogeneous and a great part of them are borderline neuroendocrine carcinomas falling between typical SCLC and LCNEC. On account of accumulated knowledge, we speculated that immunogenomically characterizing SCLC and LCNEC collectively as one group, or rather morphologically or cytologically separating SCLC from LCNEC has superior clinical value.
a9ded1e5ce5d75814730bb4caaf49419 Method
We analyzed gene expression profiles of 44 SCLCs, 56 LCNECs and 25 normal lung samples obtained from Gene Expression Omnibus. Unsupervised and supervised analyses were performed to understand molecular characteristics of samples. Pathway and CIBERSORT analyses were employed to obtain immune landscape of SCLC and LCNEC.
4c3880bb027f159e801041b1021e88e8 Result
Unsupervised clustering with 1189 differentially expressed genes revealed 2 distinct molecular subgroups (G1 and G2) of SCLC and LCNEC, which is not associated with histopathology. Targeted pathway analysis found that G1 was marked by activated IL-17, MAPK and Hippo signaling pathways. In contrast, transcriptional factors, such as ASCL1, INSM1, SOX2, and NKX2-1 were significantly up-regulated in G2, but not in G1. Moreover, in silico analysis of cellular composition and expression of immune genes disclosed unique immunoprofiles for G1 and G2. G1 was characterized by enriched CD4 memory cells, M1 macrophages and activated dendritic cells. While G2 was composed of high fractions of memory B cells and naïve CD4 cells. Strikingly, expression of both immunoinhibitors (IL10, PDL1, IDO1) and immunomodulators (OX40L, BAFF, GITR, IL6), as well as MHC class I and II molecules was higher in G1 compared to that in G2.
8eea62084ca7e541d918e823422bd82e Conclusion
We identified the common intrinsic features and molecular subgroups of SCLC and LCNEC, which are beyond conventional histopathology and better associated with immunogenomics of tumors. Further research is warranted to identify potential clinical implication of SCLC and LCNEC molecular subgroups.
6f8b794f3246b0c1e1780bb4d4d5dc53


