Virtual Library
Start Your Search
Yanfang Guan
Author of
-
+
MA16 - Novel Mechanisms for Molecular Profiling (ID 917)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Advanced NSCLC
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 13:30 - 15:00, Room 203 BD
-
+
MA16.06 - EGFR Clonality and Tumor Mutation Burden (TMB) by Circulating Tumor DNA (ctDNA) Sequencing in Advanced Non-Small Cell Lung Cancer (NSCLC) (ID 13146)
14:05 - 14:10 | Author(s): Yanfang Guan
- Abstract
- Presentation
Background
TKI has significantly improved survival time of NSCLC pts with sensitive mutation. However, pts present different outcome while receiving TKI treatment. We conduct a prospective multicenter clinical trial to determine whether clonality of sensitive mutation is related to the efficacy of TKI. We also evaluate the consistency of TMB between tissue and blood in this cohort.
a9ded1e5ce5d75814730bb4caaf49419 Method
Paired tumor and plasma samples at diagnosis were obtained from systemic treatment naïve pts with advanced NSCLC. DNA was sequenced by target-capture deep sequencing of 1021 previously annotated genes related to solid tumors. Clonal EGFR mutation was defined if EGFR mutation was in the cluster with the highest mean variated allele frequency with PyClone, and otherwise subclonal EGFR mutation. TMB of tissue (tTMB) and blood (bTMB) analysis interrogated single nucleotide variants, small insertion and deletion, with VAF ≥3 % and ≥0.5 %, respectively. TMB-high pts were identified with ≥9 mut/MB (upper quartile of data from geneplus).
4c3880bb027f159e801041b1021e88e8 Result
During February 2017 to April 2018, 127 advanced NSCLC pts were enrolled from 9 centers. A total of 653 somatic variations were detected in tissues. Mutations occurred most frequently in EGFR (57 %), TP53 (54 %), KRAS (9 %), ALK (8 %). In matched plasma, 405 (62 %) tumor-derived mutations were detected by pan-caner panel sequencing. A total of 90 EGFR mutations were detected in 73 pts, most of which occurred in tyrosine kinase domain (L858R, 41%; Ex19del, 33%). Most EGFR mutation were clonal in tissue and plasma, with a consistence of 83 % in paired samples. In addition, bTMB was significantly correlated to tTMB (Pearson r= 0.85, p-value= 1.8e-30), with a consistence of 89 %. Interestingly, high TMB was observed in a small fraction of patients (8 %) with driver mutations, such as mutations in EGFR, ALK fusion, ERBB2 and PIK3CA.
8eea62084ca7e541d918e823422bd82e Conclusion
Deep sequencing with the pan-cancer panel can effectively detect mutations and evaluate TMB in both tissue and blood with high consistence. EGFR mutations can be clonal or subclonal in both tissue and blood. Prospective multicenter study is ongoing to determine the EGFR clonality as a predictive factor for the TKI efficacy in NSCLC (TRACELib-NSCLC, NCT03059641).
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
P1.04 - Immunooncology (Not CME Accredited Session) (ID 936)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.04-34 - Study on Treatment of Stage IV Solid Tumors with Mutant Neoantigen Specific T Cells (ID 12833)
16:45 - 18:00 | Author(s): Yanfang Guan
- Abstract
Background
As an important tumor immunotherapy, the specificity and efficiency of PD1 inhibitor is not yet satisfactory. The treatment of solid tumor with mutant neoantigen specific T (Nas-T) cells developed in this study is an adoptive cell therapy which is specific for each patient. The aim is to explore the difference in safety and efficacy between Nas-T cells and PD1 inhibitors, and to evaluate the charateristic of immune repertoire (IR) as predictive biomarker.
a9ded1e5ce5d75814730bb4caaf49419 Method
A total number of 11 patients with advanced solid tumors who failed after multiline treatments were recruited. They were treated with Nas-T cells, PD1 inhibitors and BSC; other 11 patients were treated with PD1 inhibitors and BSC as control. Peripheral blood was collected at baseline and per cycle (21-28d) respectively. Multiple PCR and NGS on TCR beta chain was used to detect IR.
4c3880bb027f159e801041b1021e88e8 Result
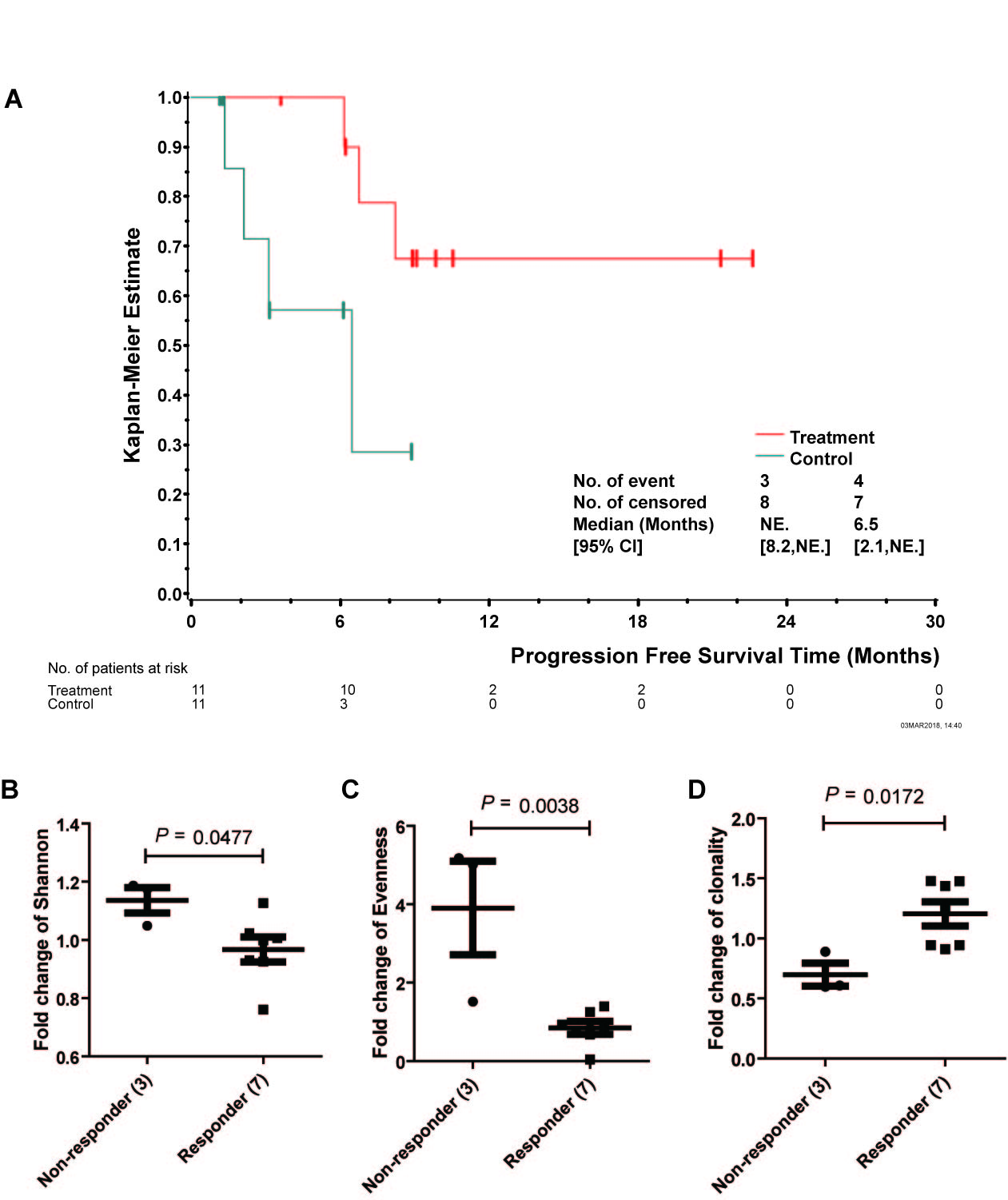
PFS of two groups had a statistical significance (P<0.05), suggesting Nas-T cells prolong patients' PFS. The safety was analyzed from routine blood urine stool test, coagulation function, liver and kidney function. There was no significant difference at baseline (P>0.05). Compared with C group, total protein and albumin in T group had a transient decrease in 3rd, 4th and 5th follow-up respectively (P<0.05), however, It can be recovered autonomously before 6th cycle.
Three indexes were examined to illuminate the diversity and clonality of IR. Compared to baseline, T cell repertoir of non-responders and responders after 1st cycle showed significant changes: Shannon 1.14 vs 0.97, P=0.048; Evenness 3.90 vs 0.85, P=0.004; Clonality 1.20 vs 0.70, P=0.017. Elevated Clonality may indicate amplification of tumor specific T cells which could recognize mutant neoantigen specifically.
8eea62084ca7e541d918e823422bd82e Conclusion
The combined immunotherapy of Nas-T cells and PD1 inhibitors is more effective than PD1 inhibitor alone in prolonging the PFS, and has a good safety. IR Clonality change shows its potential as a predictive biomarker.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P1.13 - Targeted Therapy (Not CME Accredited Session) (ID 945)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.13-18 - Exploring the Resistance Mechanism of Osimertinib and Monitoring the Treatment Response Using Plasma ctDNA in Chinese NSCLC Patients (ID 13297)
16:45 - 18:00 | Author(s): Yanfang Guan
- Abstract
Background
Osimertinib (AZD9291; Tagrisso) is a third generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) known to be effective for patients harboring the EGFR-T790M variant, which is accounts for more than half of the acquired resistance mechanisms to the first generation EGFR-TKIs. However, limited osimertinib resistance-mechanism was reported. Study on potential osimertinib-resistance mechanisms in advanced NSCLC is necessary.
a9ded1e5ce5d75814730bb4caaf49419 Method
This study enrolled eight T790M-positive (tissue validated) patients, treated with osimertinib after first generation EGFR-TKI (Erlotinib, Gefitinib, Icotinib) resistance and progressed rapidly. Serial plasma samples were collected until disease progressed. Plasma DNA was extracted and sequenced by target-capture deep sequencing of 1021 previously annotated genes related to solid tumors. Clonal EGFR T790M mutation was defined if mutation was in the cluster with the highest mean variated allele frequency with PyClone, and otherwise subclonal EGFR T790M mutation. Molecular tumor burden index (mTBI) was calculated with the mean variant allele frequency of mutations in trunk clonal population.
4c3880bb027f159e801041b1021e88e8 Result
The median progression-free survival (PFS) of these eight rapidly-progressed patients was 3.82 months [95% CI 2.05-5.01] .Targeted capture sequencing of pretreatment ctDNA showed all of the eight patients (100%) were EGFR-positive (Exon19del [n=6] and L858R [n=2]), and seven patients (88%) harbored EGFR T790M mutation, except for the only one patient (P006) who showed an extremely low level of ctDNA. During the Osimertinib treatment, five patients (63%) had osimertinib resistance-related mutations: EGFR C797S (in cis position), G724S, KRAS G12D, PIK3CA E542K, EGFR amplification, and ERBB2 amplification. Among them, two patients had more than one resistance mechanisms: patient P034 had EGFR G724S, KRAS G12D and EGFR amplification, simultaneously; patient P013 had amplification in both EGFR and ERBB2. Other potential resistance mechanisms were identified including EGFR T751I and K754E mutations in P002 and ERBB2 S603 in P013. Notably, the only one patient (P004) who had not been detected to have any known osimertinib resistance mechanism but progressed in 3 months, was demonstrated to harbor a subclonal EGFR T790M mutation by analysis of ctDNA clonal structure. Serial ctDNA monitoring showed mTBI increased when disease progressed in 88% (7/8) patients, except P006, whose mutation were negative at second (stable disease) and third (progressed disease) therapeutic evaluations due to the extremely low level of ctDNA.
8eea62084ca7e541d918e823422bd82e Conclusion
This study presented comprehensive the resistance mechanism of osimertinib progressed rapidly in ctDNA including multiple mechanisms co-occurred in same patient. Serial monitoring of plasma ctDNA may be a promising approach to explore resistance mechanism and monitored the treatment response of third generation EGFR-TKI.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P3.01 - Advanced NSCLC (Not CME Accredited Session) (ID 967)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/26/2018, 12:00 - 13:30, Exhibit Hall
-
+
P3.01-64 - Preliminary Data of Diverse Therapies in Patients with Advanced Non–Small-Cell Lung Cancer Harbouring RET-Rearrangement (ID 13677)
12:00 - 13:30 | Author(s): Yanfang Guan
- Abstract
Background
Activating RET-rearrangement has been discovered to play a crucial role in NSCLC tumorigenesis. However, the lack of specificity narrowed efficacy of multi-kinase inhibitors (MKIs) and the optimal treatment remains unknown. In this study, we compared chemotherapy, immunotherapy and MKIs in this group of patients.
a9ded1e5ce5d75814730bb4caaf49419 Method
We retrospectively evaluated the efficacy of these three treatments in advanced, RET-rearranged NSCLC patients between January 2013 and April 2018 at our institution. RET-rearrangements were assessed by Next-generation sequencing (NGS) or any of FISH, IHC, RT-PCR. Treatment data were collected after the patients had been diagnosed with RET-rearranged advanced NSCLC. Progression-free survival (PFS) was measured from treatment start to disease progression, all-cause mortality or last follow up. Median follow-up time was 5.1months. NGS was performed to assess somatic mutation of available samples.
4c3880bb027f159e801041b1021e88e8 Result
A total of 30 patients with RET-rearrangement were investigated in this study. After the diagnosis, 15 patients, genetic profiles confirmed by NGS, received chemotherapy (n=10), checkpoint-inhibitors (n=7) and RET targeted MKI (n=6) with evaluable response. Several patients take any two of these three treatments as different line therapies. The disease control rate of chemotherapy, immunotherapy, MKI group was 70.0%, 71.43% and 50%, respectively. While the median PFS of three groups was 2.50 months, 2.70 months, 0.30 months, respectively, which of no significance. The NGS data of 10 patients showed that RET-rearrangement co-occurred with several other genes, including TP53, NTRK, CDK4, ERBB4. A low mutation burden (mean 4.5 mutations) was observed (Figure 1).
8eea62084ca7e541d918e823422bd82e Conclusion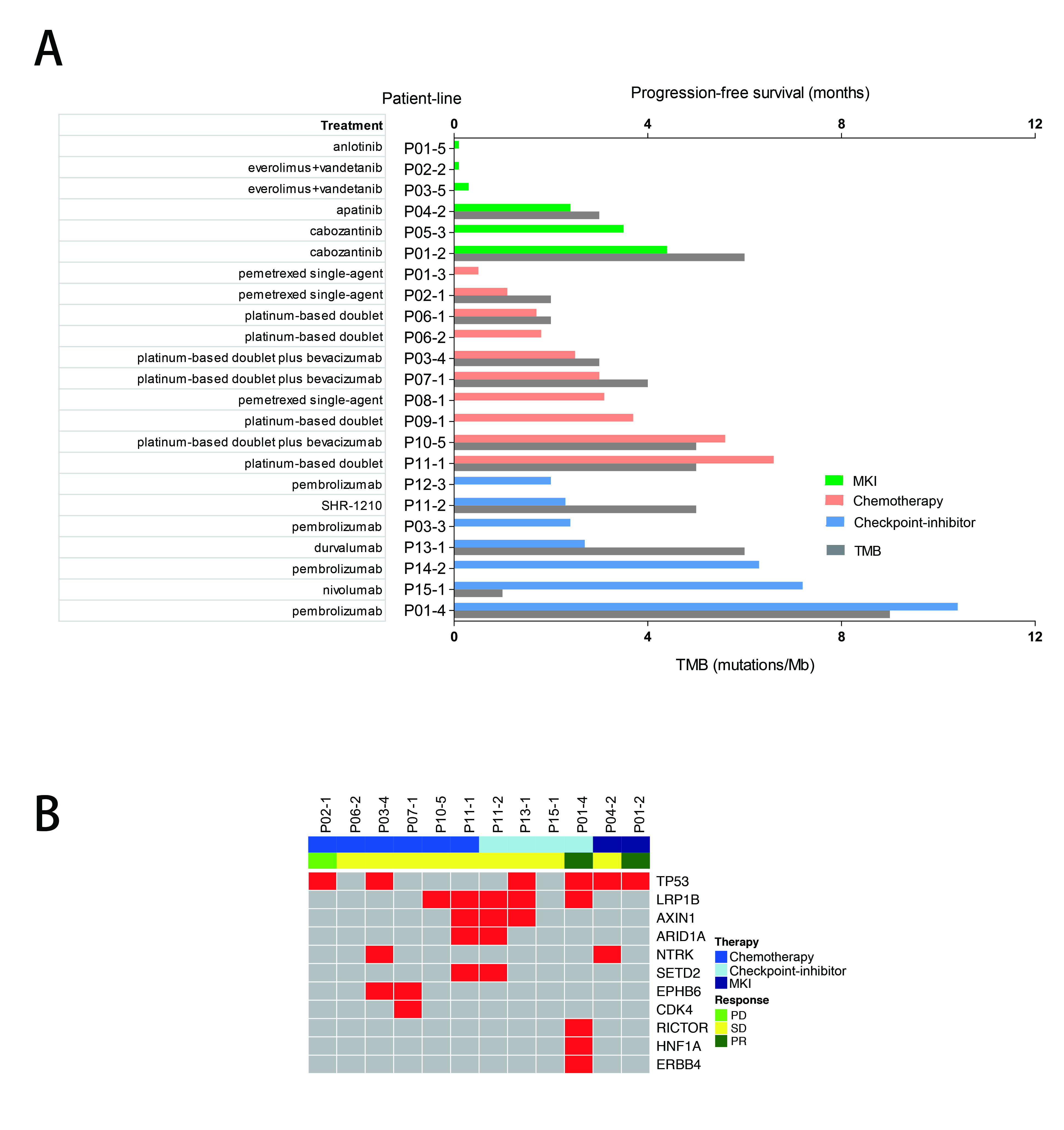
We confirmed relatively low PFS in advanced RET-rearranged NSCLC with MKIs reported in previous studies. But further investigation is warranted. Treatment with checkpoint-inhibitors seemed to encouragingly prolong PFS but a larger group of patients is needed to draw a definite conclusion.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P3.03 - Biology (Not CME Accredited Session) (ID 969)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/26/2018, 12:00 - 13:30, Exhibit Hall
-
+
P3.03-01 - BRAF V600 and Non-V600 Mutations in Chinese Lung Cancer (ID 13759)
12:00 - 13:30 | Author(s): Yanfang Guan
- Abstract
Background
BRAF gene mutation, especially V600E, was frequently mutated in cancer. Vemurafenib and dabrafenib has already been approved in melanoma as well as NSCLC and preclinical studies have demonstrated promising results in non-V600 NSCLC. But the landscape of BRAF non-V600 mutation in Chinsese lung cancer was rarely descripted.
a9ded1e5ce5d75814730bb4caaf49419 Method
7,417 normal-paired samples from lung cancer patients were analyzed using hybridization capture-based next generation sequencing and alterations including single nucleotide variants (SNVs), short insertions/deletions (indels), copy number variations (CNVs) and structural variations (SV) were analyzed.
4c3880bb027f159e801041b1021e88e8 Result
BRAF was altered in 1.8% (133 of 7,417) of all tumors. BRAF V600 (gain of function, GOF) and non-V600 mutations (GOF: G469V/R/E/A, K601N/E, L597V/R, T599dup/T599R, N486_P490del, L525R, and loss of function/LOF: D594N/G, N581S/I, G466V/A, K483E, G596R), has previously been reported to increase MEK/ERK activation, were detected in 52.3% (58/111) and 47.7% (53/111) of BRAF functional mutation patients. CNVs and SVs were both observed at a frequency of 0.9% (1/111). Two patients have two GOF mutations (V600E/T599R). We also found that 82.0% (91/111) of the BRAF functional mutation carriers also owned the other actionable or driver mutation, the most frequent one was TP53 (68.1%), then was EGFR (18.7%), KRAS/NRAS (14.3%), PIK3CA (11.0%) as well as CDKN2A/B (9.9%).
8eea62084ca7e541d918e823422bd82e Conclusion
BRAF gene mutations, non-V600 especially, was extensively mutated in Chinese lung cancer. This work supports a broad profiling approach in lung cancers and suggests that non-V600E BRAF alterations represent a subgroup of lung cancers in which targeted therapy should be considered.
6f8b794f3246b0c1e1780bb4d4d5dc53


