Virtual Library
Start Your Search
Jun-Yi Ye
Author of
-
+
MA15 - Colliding Approaches - EGFR and Immunotherapy (ID 916)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Targeted Therapy
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 13:30 - 15:00, Room 107
-
+
MA15.06 - Circulating Tumor DNA Portrays the Resistance Landscape to a Novel Third Generation EGFR Inhibitor, AC0010 (ID 13641)
14:05 - 14:10 | Author(s): Jun-Yi Ye
- Abstract
- Presentation
Background
In a Phase I/II dose-escalation and expansion study conducted at Guangdong Lung Cancer Institute, AC0010 demonstrated promising efficacy and good tolerability in advanced NSCLC patients with EGFR T790M-mediated resistance to previous EGFR TKIs, (NCT02330367). As disease progression (PD) with EGFR T790M-directed therapy also emerges over time, we investigated the resistance mechanisms to AC0010 in this study.
a9ded1e5ce5d75814730bb4caaf49419 Method
Serial ctDNA samples obtained from patients who developed PD with AC0010 were analyzed using ultra-deep sequencing capturing 295 cancer-related genes. Alterations that were absent before treatment and acquired at PD or that increased in abundance during treatment were identified as putative resistance mechanisms.
4c3880bb027f159e801041b1021e88e8 Result
Longitudinal plasma samples were obtained from 23 patients who progressed on AC0010 (data cut-off October 14, 2016; figure1). Putative resistance mechanisms to AC0010 were identified in 19/23 patients (>1 putative resistance mechanism was detected in some patients). EGFR amplification was the predominant resistance mechanism (21.1% [4/19 patients]), followed by TP53 loss of heterozygosity (15.8% [3/19]). EGFR C797S mutation, Met amplification and mutations in the PI3KCA pathway each occurred in 10.5% of patients (2/19). SCLC transformation, ERBB2 amplification, CD79A_A32G mutation, CDKN2A_R80 mutation, CRLF2 amplification, MLH1 amplification, Rb1 loss, and concurrent rise in the allelic fraction of tumor suppressor gene TP53 and Rb1 were each detected in 5.3% of patients (1/19). In a patient with PD following single-agent AC0010 and EGFR amplification as the putative resistance mechanism to AC0010, subsequent treatment with AC0010 plus nimotuzumab (EGFR monoclonal antibody) successfully overcame resistance, resulting in a response that lasted for 7.7 months.
8eea62084ca7e541d918e823422bd82e Conclusion
The resistance landscape to AC0010 appears to differ from that described previously with osimertinib. In this cohort of patients in China, EGFR amplification was the predominant resistance mechanism to AC0010 and could be potentially overcome by EGFR dual inhibition.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
MA16 - Novel Mechanisms for Molecular Profiling (ID 917)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Advanced NSCLC
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 13:30 - 15:00, Room 203 BD
-
+
MA16.10 - Clinical Utility of Cerebrospinal Fluid Cell-Free DNA for Clarifying Genetic Features of Leptomeningeal Metastases in ALK Rearrangement NSCLC (ID 12142)
14:35 - 14:40 | Author(s): Jun-Yi Ye
- Abstract
- Presentation
Background
Leptomeningeal metastases (LM) were associated with a poor prognosis in non small cell lung cancer (NSCLC). LM were much more frequent in EGFR mutant patients, and cerebrospinal fluid (CSF) cell-free DNA (cfDNA) has shown unique genetic profiles of LM in patients harboring EGFR mutations in our previous studies. However, studies in ALK positive NSCLC patients with LM are scarce.
a9ded1e5ce5d75814730bb4caaf49419 Method
Lung cancer patients with ALK rearrangement were screened from Sept 2011 to Feb 2018 at our institute. Leptomeningeal metastases were diagnosed by MRI or CSF cytology or next-generation sequencing (NGS) of CSF cfDNAs. Paired plasma were also tested by NGS.
4c3880bb027f159e801041b1021e88e8 Result
LM were diagnosed in 22 (7.6%) of 288 ALK rearrangement patients with lung cancer. A total of 11 ALK positive patients with LM were enrolled with CSF cfDNA tested by NGS (one case used CSF precipitates instead of CSF cfDNA). Paired plasma were available in 11 patients. Driver genes were detected in 75.0% CSF samples and 45.5% plasma respectively (P=0.214). Max allele fractions were higher in CSF cfDNA than in plasma (40.8% versus 0%, P=0.021). ALK variant 1 (E13:A20) was detected in 3 cases of CSF and paired plasma, respectively. ALK variant 2 (E20:A20) was identified in 5 cases of CSF and 1 paired plasma. Multiple copy number variants (CNV) were mainly found in CSF cfDNA, including EGFR copy number gains. Resistance mutations including gatekeeper gene ALK G1202R was identified in CSF cfDNA with ALK variant 1 and ALK G1269A was detected in plasma. The detection rate of TP53 was 45.4% versus 27.3% in CSF cfDNA and plasma.
8eea62084ca7e541d918e823422bd82e Conclusion
CSF cfDNA was more sensitive than plasma to reveal genetic features of ALK-fusion LM, confirming its role as a liquid biopsy medium for LM in driver gene positive NSCLC.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
MA21 - Molecular Subtyping, CBL3, and Non Coding RNA (ID 924)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Biology
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 15:15 - 16:45, Room 205 BD
-
+
MA21.05 - Comprehensive Genomic Characterization and Prognostic Nomogram Developed by 295-Gene Panel Targeted Sequencing (ID 13664)
15:45 - 15:50 | Author(s): Jun-Yi Ye
- Abstract
- Presentation
Background
The genomic landscape of lung cancer has been thoroughly studied in the western population. But comprehensive genetic profiling reports have been limited for the Chinese patients. Here we conducted deep targeted sequencing on a large cohort of Chinese treatment-naïve lung cancer patients and identified novel molecular patterns. We developed nomogram models for prognosis prediction by integrating genetic and clinical characteristics and aim to explore more precise models for risk stratification beyond TNM staging.
a9ded1e5ce5d75814730bb4caaf49419 Method
This was a retrospective study, enrolling diagnosed lung cancer patients at Tianjin Medical University Cancer Institute & Hospital from 2009 to 2012. We developed genomic landscape by targeted sequencing using a panel consisting of exons and critical introns of 295 cancer-related genes. Nomogram models were established to provide risk stratification in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) patients
4c3880bb027f159e801041b1021e88e8 Result
513 tumor tissue samples were collected at very beginning of treatment (stage I, n=193; stage II, n=140; stage IIIA, n=140; stage IIIB, n=5; stage IV, n=28; unknown, n=7). The most frequently mutated oncogenic genes in LUAD were EGFR (55%) and KRAS (11%), compared to 14% and 33% in TCGA. Heterogeneity existed in terms of mutual exclusive and co-occurrent mutated gene pairs between LUAD and LUSC. In LUAD, pairs with most significant exclusivity was EGFR/KRAS and co-occurrent was NOTCH3/GRIN2A, whereas the most significant concurrent gene pair in LUSC was ZNF703/FGFR1. To predict survival, our nomograms identified that, in stage I-IIIA LUAD and LUSC, mutated TET2 contributed to more favorable DFS while mutations in EPHA3 and ETV5 indicated better OS. Stage and mutated KRAS were associated with inferior DFS and OS (DFS, n=222, c-index=0.714; OS, n=308, c-index=0.706). In the T1+2&N0&M0 subgroup, which is considered clinically low risk for relapse, older patients who carry BRCA2 mutations were found to strongly correlate with poor DFS (n=121, c-index=0.709), while age and mutated KRAS were distinct indicators of inferior OS (n=163, c-index=0.725). The calibration for survival probability displayed well agreement between nomogram prediction and actual outcomes. Stratification of different risk groups based on nomogram prediction displayed significant differences among Kaplan-Meier curves for survival outcomes (p<0.0001).
8eea62084ca7e541d918e823422bd82e Conclusion
This is the so far largest cohort of Chinese lung cancer patients with comprehensive genomic profiling reported. We revealed unique molecular profiles than TCGA and distinct mutual exclusivity and co-occurrence patterns between LUAD and LUSC. In addition, the nomogram models show promise of more precise prognostic prediction of NSCLC patients when integrating genetic information with clinical characteristics.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
P1.01 - Advanced NSCLC (Not CME Accredited Session) (ID 933)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.01-55 - Unique Genetic Profiles from Cerebrospinal Fluid Could Predict Survival of EGFR-Mutant NSCLC with Leptomeningeal Metastases (ID 12369)
16:45 - 18:00 | Author(s): Jun-Yi Ye
- Abstract
Background
Leptomeningeal metastases (LM) are more frequent in NSCLC with EGFR mutations;and cerebrospinal fluid (CSF) could reveal the unique genetic profiles of LM in our previous studies, but whether they could predict the overall survival (OS) of LM remains unknown.
a9ded1e5ce5d75814730bb4caaf49419 Method
EGFR-mutant NSCLC patients with LM were enrolled,and clinical data and genetic profiles detected by Next-generation sequencing were collected. We further drew nomogram with endpoint of OS after LM, then performed index of concordance (C-index) and survival analysis to evaluate predictive role.
4c3880bb027f159e801041b1021e88e8 Result
In total, 61 patients were enrolled and all with genetic profiles from CSF. Patents with high copy number variations (CNVs) or harboring CDK6, TP53 exon5 or FGF19 in CSF demonstrated significant poorer OS than those without (Fig. 1). Cox regression analysis indicated CNVs, CDK6,CDKN2A,TP53,MET and NTRK1 as prognostic factors and further selected for nomogram (Fig. 2). C-index of nomogram was 0.743, indicating the moderate predictive effect. In the calibration curves, we scored the patients based on the model, using bisection and trisection methods to divide into low and high points groups; and low, medium and high points groups (Fig. 3), and significant difference were found in both the survival analyses (NA versus 7.47months, P<0.01) and (NA, 10.33 versus 4.43 months, P<0.01) respectively. Patients who received Osimertinib after LM seemed to have longer OS than those who did not (14.5 months versus 7.7 months) but without significant difference(P=0.10); however interestingly, in those with EGFR T790M negative who took Osimertinib after LM by themselves obtained survival benefit than those who did not(NA versus 7.7 months, P=0.04), and the results needed to be validated.
8eea62084ca7e541d918e823422bd82e Conclusion
Unique genetic profiles from CSF could well predict OS of LM. High CNVs, CDK6, TP53 exon5 or FGF19 in CSF in CSF may be related to poor prognosis of LM.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P1.03 - Biology (Not CME Accredited Session) (ID 935)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.03-34 - Combined Molecular and Radiological Evaluation Unveils Three Subtypes of Disease Progression to a Third Generation EGFR TKI (ID 12055)
16:45 - 18:00 | Author(s): Jun-Yi Ye
- Abstract
Background
The definition of disease progression (PD) to EGFR TKIs has evolved from RECIST to a combination of clinical and RECIST evaluation. Patients with dramatic, local or gradual progression to third generation EGFR TKIs have been tailored to different subsequent treatment strategies. However, little is known about progression to third generation EGFR TKIs from molecular perspective.
a9ded1e5ce5d75814730bb4caaf49419 Method
Longitudinal plasma samples were collected from T790M-positive patients who progressed on a third generation EGFR TKI AC0010 in a phase I/II study in Guangdong Lung Cancer Institute. A pre-defined and unified molecular and radiological evaluation of PD were performed. Ultra-deep sequencing capturing 295 cancer-related genes was performed to track the changes in ctDNA to depict molecular PD, which was defined by acquired SNV/SCNV, or ≥20% increase in allelic fraction/copy number of pre-existing SNV /SCNV or both. Radiological PD was defined by RECIST.
4c3880bb027f159e801041b1021e88e8 Result
As of October 2016, 102 serial plasma samples from 23 patients with clinical PD were included. Three subtypes of PD to AC0010 were revealed (Fig1). Molecular PD occurred prior to radiological PD in 43.5% of patients (10/23), with an average lead time of 3.0 months. Molecular PD occurred concurrently with radiological PD in 39.1% of patients (9/23). Interestingly, 17.4% of patients (4/23) experienced radiological PD prior to molecular PD, with molecular PD occurred during AC0010 continuation beyond progression (CBPD) in 3 patients. Of patients experienced clinical stable PD in extracranial lesions, radiological PD occurring prior to molecular PD group (n=2) demonstrated longer duration of AC0010 CBPD than molecular PD occurring prior to (n=3) or concurrently with radiological PD groups (n=4) (Median, 5.6 months vs. 1.9 months vs. 1.8 months).
8eea62084ca7e541d918e823422bd82e Conclusion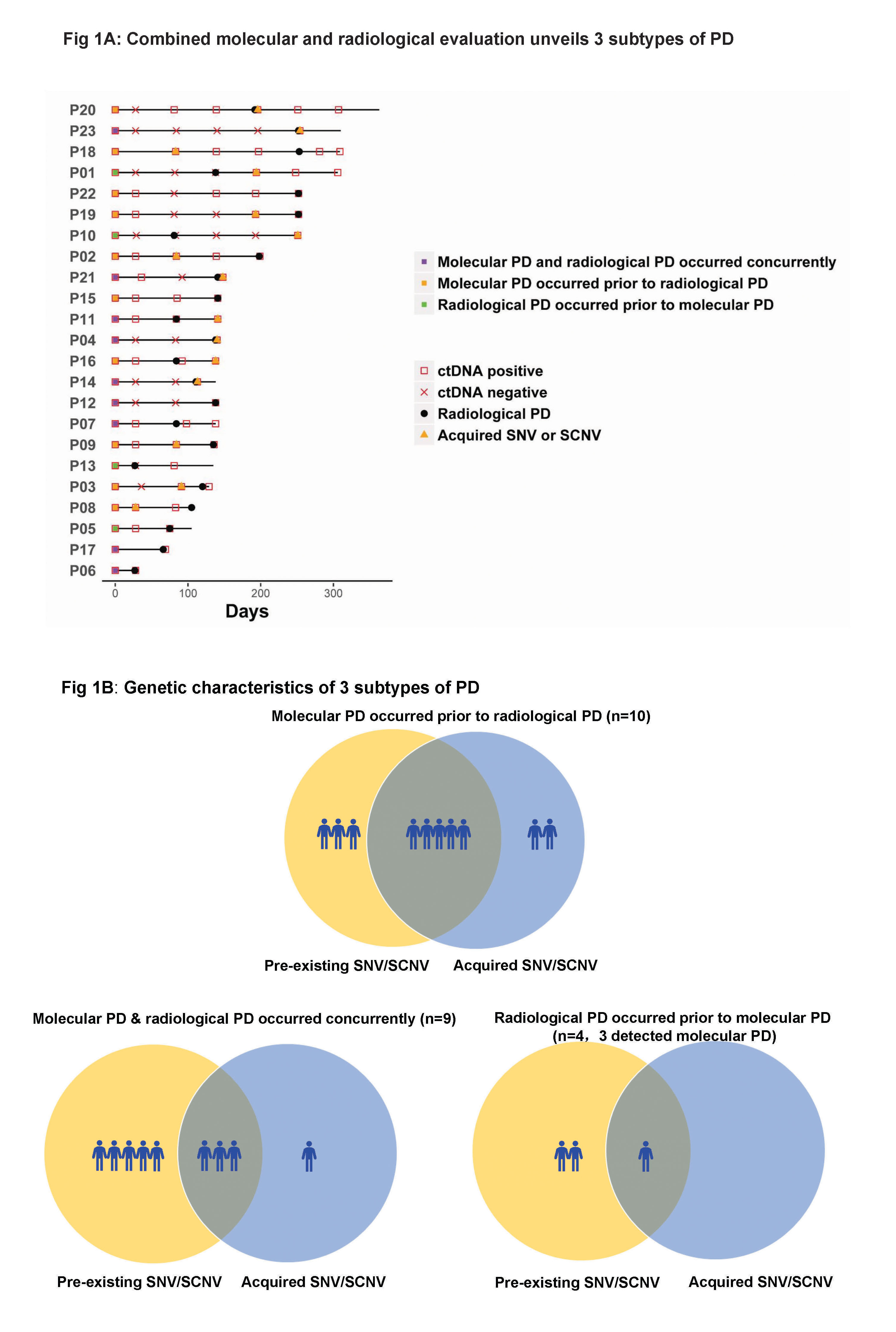
Our study revealed 3 distinct subtypes of PD to AC0010, providing insights into PD by combining molecular and radiological evaluation and might guide the optimal time for treatment switch and personalized subsequent treatments.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P2.01 - Advanced NSCLC (Not CME Accredited Session) (ID 950)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 16:45 - 18:00, Exhibit Hall
-
+
P2.01-44 - Prognostic Value of TP53 Hot Exon Mutation in Patients with Advanced Non-Small Cell Lung Cancer (NSCLC) (ID 13150)
16:45 - 18:00 | Author(s): Jun-Yi Ye
- Abstract
Background
Numerous studies have revealed either very marginal or no prognostic value of TP53 mutation NSCLC patients. Currently, in clinical settings, all TP53 mutations have been considered equally, without any differentiation between the various types and positions of mutations. However, increasing evidence has triggered us to challenge such practice.
a9ded1e5ce5d75814730bb4caaf49419 Method
We retrospectively investigated the correlation between mutations occurring at hot exons (5-8) and overall survival (OS) in 214 previously tyrosine kinase inhibitor (TKI) treated advanced NSCLC patients. Among them, 184 had 1 line of TKI-treatment and the remaining 30 patients had more than 1 line of TKI-treatment. 115 harbored TP53 mutation; among them 105 patients had concurrent EGFR mutation; 5 had ALK rearrangements; 1 had ROS1 rearrangements; 1 had KRAS and 2 had ERBB2 mutations. 99 patients had wild type (WT) TP53; among them, 92 had EGFR mutation, 4 with ALK-rearrangements, 1 with MET and 1 with BRAF mutation. Fisher’s exact test and the Mann-Whitney test were used to determine if categorical and continuous variables, respectively, differed between TP53 WT and mutant groups.
4c3880bb027f159e801041b1021e88e8 Result
The prevalence of TP53 mutation in our cohort is 53.7% (115/214); 28 had mutation on exon 5, 18 on exon 6, 22 on exon 7 and 32 on exon 8. 32 patients had loss of function mutation and 51 patients had disruptive mutation. Our data revealed a positive correlation with N and M stage. Patients harboring TP53 mutation are more likely to diagnose with more advanced N (p=0.018) and M stage (p=0.001). Furthermore, patients with TP53 mutation are more likely to have liver (p<0.001) and bone metastasis (p=0.012). In patients treated with only 1 line of TKI-treatment, although TP53 status had no effect on PFS (p=0.241) and OS (p=0.49) when they were considered collectively, we observed patients with mutation in exon 5 had shorter OS (p=0.029) and mutation in exon 8 had shorter PFS (P=0.003) after controlling for age, gender, stage and histology. Furthermore, within the osimertinib subgroup (N=101), patients harboring mutation in exon 8 had significantly shorter PFS (P=0.007). In patients treated with more than 1 line of treatment, neither TP53 mutation considered collectively, nor hot exon mutations had correlation with PFS or OS.
8eea62084ca7e541d918e823422bd82e Conclusion
Our study revealed unfavorable prognostic value of mutations in exon 5 and no prognostic value of TP53 if all mutations were considered collectively. Our study adds new dimension to the emerging picture that not all TP53 mutants are equal.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P3.12 - Small Cell Lung Cancer/NET (Not CME Accredited Session) (ID 978)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/26/2018, 12:00 - 13:30, Exhibit Hall
-
+
P3.12-12 - Genomic Profiling of Pulmonary Large-Cell Neuroendocrine Carcinoma (LCNEC) Reveals Distinct Mutational Landscape (ID 13244)
12:00 - 13:30 | Author(s): Jun-Yi Ye
- Abstract
Background
The controversial classification of lung neuroendocrine tumor has been amended a few times since recognised as a separate entity. LCNEC shares clinical features with small cell lung carcinoma (SCLC) and they were both classified as lung neuroendocrine carcinoma according to the 2015 WHO lung primary pathology classification, numerous studies have revealed barely satisfactory outcomes when it was treated as SCLC. However the underlying molecular basis for such commonalities and discrepancies are poorly understood. In this study, we interrogated the genomic landscape of LCNEC and SCLC along with their histologically related subtypes: carcinoids and atypical carcinoids to define the molecular pattern of LCNEC.
a9ded1e5ce5d75814730bb4caaf49419 Method
We performed targeted sequencing in 35 tissue samples using a panel covering 520 cancer related genes, spanning 1.6MB of human genome, with an average sequencing depth of 1,418x. Among them, 15 were diagnosed with SCLC, 9 with LCNEC, 6 with carcinoid and 5 with atypical carcinoid.
4c3880bb027f159e801041b1021e88e8 Result
On average, LCNEC exhibited 13.5 mutations per million base pairs (Mb) and a C:G>A:T transversion rate of 34%, which is indicative of tobacco exposure. LCNEC had SCLC (16.7 Mb) had comparable TMB (p=0.18), which is significantly higher than carcinoids (1.2/Mb, p<0.001) and atypical carcinoids (2.4/Mb, p<0.001). The most frequently mutated gene in LCNEC is TP53 (89%, 8/9), followed by NOTCH1 (33%), KEAP1 (22%), RB1 (22%) and a few chromatin modifiers, including KMT2D (33%), KMT2C (33%). Co-mutation in TP53 and RB1, a hallmark of SCLC, was found in 22% (2/9) of LCNEC patients; in contrast, 80% of SCLC patients harbored concurrent mutation. 67% carcinoid (4/6) and 20% (1/5) atypical carcinoid patients had no mutation identified from this panel. No classic lung adenocarcinoma driver mutations were found in any subtype. Copy number analyses revealed significantly higher copy number variation (CNV) in SCLC and LCNEC comparing with carcinoids and atypical carcinoids, which yield virtually no CNV. Our analysis revealed a comparable CNV status of SCLC and LCNEC (p=0.158), with an enrichment in amplification of chromatin modifiers.
8eea62084ca7e541d918e823422bd82e Conclusion
Our study, comprehensively characterized 4 subtypes of neuroendocrine tumors, revealed a high TMB and CG:AT transversion rate in LCNEC patients as well as a distinctive mutation landscape, with an enrichment of mutations occurring at chromatin remodelers. Furthermore, LCNEC has comparable TMB and CNV status as SCLC, which are significantly higher than carcinoid and atypical carcinoids.
6f8b794f3246b0c1e1780bb4d4d5dc53


