Virtual Library
Start Your Search
Victor Cohen
Author of
-
+
MA09 - Lung Cancer Surgical and Molecular Pathology (ID 908)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Pathology
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 15:15 - 16:45, Room 202 BD
-
+
MA09.09 - EBUS-TBNA in Assessing PD-L1 Expression in NSCLC (ID 13471)
16:15 - 16:20 | Author(s): Victor Cohen
- Abstract
- Presentation
Background
a9ded1e5ce5d75814730bb4caaf49419 Method
Pembrolizumab is the only immunotherapy approved as a first line agent for metastatic NSCLC in patients with high programmed death‐ligand 1 (PD‐L1) expression. The standard samples for PD-L1 testing are considered surgical or core biopsies. In this study, our primary objective is to identify the adequacy of endobronchial ultrasound-guided transbronchial needle aspiration (EBUS TBNA) tumor samples in detecting PD-L1 expression.
Between July 2016 and April 2017 a total of 1352 consecutive cases of non-small cell lung cancer (NSCLC) were identified. 29 specimens were deemed inadequate (less than 100 viable tumor cells) and were excluded. 1323 specimens analyzed included surgical samples (N=238), small biopsy (N=744) and cytology cell blocks (N=341). Cytology cell blocks were from EBUS-TBNA (N=190), fine needle aspiration (FNA) (N=61) and pleural/pericardial fluid (N=90). PD-L1 expression was examined by staining with Dako PD-L1 IHC 22C3 pharmDx kit. A Tumor Proportion Score (TPS) was categorized as <1%, 1-49% and ≥ 50% tumor cells.
4c3880bb027f159e801041b1021e88e8 Result
Most of the 1323 specimens (84%) were non-squamous carcinomas. Overall yield for TPS > 50% was 36%. Rate of PD-L1 positivity was no different in non-squamous (37%) compared to squamous (32%). Diagnostic yield of PD-L1 for different sample types varied substantially (Table 1). The EBUS-TBNA samples had the highest yield for TPS ≥ 50% (p=0.025).
8eea62084ca7e541d918e823422bd82e ConclusionTPS Surgical resection Small biopsy EBUS-TBNA FNA Fluid cytology Total Adequacy 100% 99% 98% 96% 92% 98% ≥ 50% 69 (29) 269 (36) 84 (44) 21 (34) 38 (42) 481 1-49% 87 (37) 274 (37) 57 (30) 22 (36) 22 (24) 462 <1% 82 (35) 201 (27) 49 (26) 18 (30) 30 (33) 380 Total 238 744 190 61 90 1323
Our results show that cytology cell blocks could be considered as a valuable resource for PD-L1 testing in advanced NSCLC. Future studies are warranted to explore clinical correlation of PD-L1 on EBUS-TBNA samples and immunotherapy outcome.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
MA21 - Molecular Subtyping, CBL3, and Non Coding RNA (ID 924)
- Event: WCLC 2018
- Type: Mini Oral Abstract Session
- Track: Biology
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 15:15 - 16:45, Room 205 BD
-
+
MA21.09 - Differential Gene Expression in Tumor and Normal Tissue Reveals New Insights in the Biology of Non-Small Cell Lung Carcinoma. (ID 13341)
16:15 - 16:20 | Author(s): Victor Cohen
- Abstract
- Presentation
Background
Effective use of targeted cancer therapies typically depends upon the identification of actionable genomics somatic alterations, benefiting only a minority of Non-Small Cell Lung Carcinoma (NSCLC) patients. To integrate transcriptomic assessment in cancer precision medicine, we have evaluated the mRNA expression levels in tumors and their matched normal lung tissues with the hypothesis that mRNA quantification in tumors relative to their matched normal tissue may better reveal small transcriptional differences that are associated with major biological effects.
a9ded1e5ce5d75814730bb4caaf49419 Method
The discovery set used 123 frozen macrodissected treament-naive NSCLC tumors and matched normal tissues from surgical resections performed at the Mutualiste Montsouris Institute (Paris, France), and the validation set used 143 FFPE macrodissected treatment-naive NSCLC tumors and matched normal tissues from surgical resections performed at the Jewish General Hospital (Montreal, QC, Canada). A pathology review was performed in all cases. In the discovery set, expression levels of 17,318 genes were analysed using an Agilent Technologies platform; in the validation set, the NanoString nCoutner technology was used with a customized 148 probeset that was designed according to the results of the discovery phase. The primary objective of the study was post-surgery progression-free survival (PFS). The secondary objectives were post-surgery overall survival (OS) and the identification of pathway-driven expression signatures.
4c3880bb027f159e801041b1021e88e8 Result
A set of highly expressed genes correlated with post-surgery PFS. Details of the prognostic signature will be presented at the meeting. Importantly, mRNA levels in normal tissues were highly variable between individuals. Organ matched reference enabled to control for the noise signals related to individual background genetic variability. The cell cycle G2/M checkpoint was the most significantly deregulated expression pathway in this cohort; nine genes in the signature are involved in this pathway and were upregulated in tumors, dependending on their histology: CHEK1, TOP2A, AURKA, CDC2, PLK1, CDC2, CDC25A, CDC25B, and CDC25C. CHEK1 is a pivotal gene in regulating the G2/M cell cycle pathway that triggers the double-strand base excision repair in which the main effector is PARP1. CHEK1 was overexpressed in 86% of adenocarcinomas, versus 42% for PARP1.
8eea62084ca7e541d918e823422bd82e Conclusion
Conventional transcriptomic approaches using expression metrics obtained in tumor pools may miss important changes due to individual variability in non-tumoral tissue.The present work illustrates that paired matched tumor and normal tissues can identify new key genes involved in the biology and pathogenesis of NSCLC, and opens new avenues for integrating transcriptomic investigations in the precision medicine arena.
6f8b794f3246b0c1e1780bb4d4d5dc53Only Members that have purchased this event or have registered via an access code will be able to view this content. To view this presentation, please login, select "Add to Cart" and proceed to checkout. If you would like to become a member of IASLC, please click here.
-
+
P1.04 - Immunooncology (Not CME Accredited Session) (ID 936)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/24/2018, 16:45 - 18:00, Exhibit Hall
-
+
P1.04-16 - Comparison of Clinical Response to Checkpoint Inhibitors in Advanced NSCLC with High PD-L1 Expression Tested on Cytology Versus Biopsy Samples (ID 12525)
16:45 - 18:00 | Author(s): Victor Cohen
- Abstract
Background
PD-L1 immunohistochemistry (IHC) expression correlates with clinical response to checkpoint inhibitors in advanced-stage NSCLC. PD-L1 IHC testing is usually performed on tissue blocks from core needle biopsy or surgical resection, but appears to be feasible on cytology cell blocks as well. In this retrospective study, we assessed the clinical response to checkpoint inhibitors in patients with NSCLC and high PDL1 IHC expression on cytology specimens in comparison with tissue biopsy specimens.
a9ded1e5ce5d75814730bb4caaf49419 Method
Between August 2015 and April 2018, 116 patients with NSCLC received immunotherapy at our institution. Only cases with known PD-L1 expression from IHC testing performed on small biopsies or cytology cell blocks were included. A total of 65 consecutive cases were reviewed, including 40 small biopsies and 25 cytology samples. A Tumor Proportion Score (TPS) was categorized as high (≥ 50% tumor cell staining) or low (<50%). Response to treatment was categorized as disease control (DC, including complete and partial response and stable disease) or progression (PD). The primary outcome was the rate of disease control.
4c3880bb027f159e801041b1021e88e8 Result
Patients were mostly current or ex-smokers (91%), Caucasians (82%) and non-squamous carcinomas (85 %). High TPS was seen in 44 (68%) cases. Immunotherapy was given in the first line setting in 20 (31%) patients, the second line in 36 (55%), and the third line in 9 (14%). 50 (77%) patients received Pembrolizumab, 10 (15%) Nivolumab and 5 (8%) others received immunotherapies on RCTs. Overall, there was DC in 40 (62%) patients and PD in 17 (26%). There was no significant difference in DC rate between the cytology and the small biopsy groups in high TPS group.
8eea62084ca7e541d918e823422bd82e Conclusion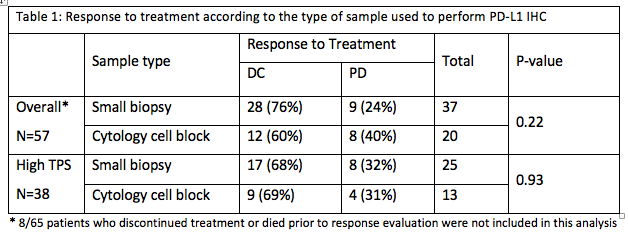
PD-L1 expression on cytology cell blocks and on small biopsies appears to have similar clinical significance. Further prospective trials are needed to confirm these findings.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P2.01 - Advanced NSCLC (Not CME Accredited Session) (ID 950)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 16:45 - 18:00, Exhibit Hall
-
+
P2.01-23 - Baseline Plasma Biomarkers Predict Long-Term Responses to ALK-TKIs in ALK+ Advanced Non-Small Cell Lung Cancer (NSCLC) (ID 13458)
16:45 - 18:00 | Author(s): Victor Cohen
- Abstract
Background
Median progression free survival (PFS) for ALK tyrosine kinase inhibitors (ALK-TKIs) range from 10.9-25.7 months (mos)[1],[2],[3],[4]. While most ALK+ NSCLC patients develop resistance to ALK-TKIs within 1-2 years, a subset of patients experience a response >2 years. Given the lack of proteogenomic markers predicting long-term responses to ALK-TKIs, we conducted whole-exome sequencing (WES) and proteomic profiling to define molecular determinants that could predict a long-term ALK-TKI response, help guide the sequentiality of therapies and optimize treatment strategies.
[1]Soria, J.C. et al. Lancet389,917–929 (2017)
[2]Peters, S. et al. N. Engl. J. Med.377, 829-838(2017)
[3]Kim, D.-W. et al.J. Clin. Oncol.35,2490–2498 (2017)
[4]Solomon, B. J. et al.N. Engl. J. Med.371,2167–2177 (2014).
a9ded1e5ce5d75814730bb4caaf49419 Method
Twenty-four patients with advanced ALK+ NSCLC were enrolled in our study. WES was performed on primary and post-treatment metastatic tissue to identify genomic aberrations and ALK fusions. MRM-MS was used to analyze 327 protein candidates in plasma collected from patients at baseline.
4c3880bb027f159e801041b1021e88e8 Result
Patients were categorized into 3 groups based on duration of response: long-term responders [LR; PFS ≥24 mos (n=8)], normal responders [R; 3 < PFS < 24 mos (n=10)] and non-responders [NR;PFS <3 mos (n=6)]. At data cutoff (30 April 2018), median PFS was 1.6 mos for NR, 11.7 mos for R and 37.1 mos for LR. Two LR remain on treatment and have experienced a PFS > 37.4 mos. Despite detecting novel ALK fusion partners, multiple somatic mutations and copy number aberrations by WES, we could not define a genomic signature predictive of a long-term response to ALK-TKIs from our small cohort. However, MRM-MS identified 15 proteins differentially regulated between LR and NR, including SODE, F13A, LYAM1, FCGBP, PGBM and LUM. Differences in protein levels were further pronounced between LR and NR. To determine whether our protein signature can discriminate according to response groups, we performed principal component and hierarchical clustering analyses. Both analyses successfully segregated LR from R and NR. Moreover, we used our set of 15 proteins to generate single-sample Gene Set Enrichment Analysis scores which distinguished LR from R and NR as distinct groups.
8eea62084ca7e541d918e823422bd82e Conclusion
Targeted proteomic profiling of baseline plasma from ALK+ NSCLC patients identified a protein signature that may predict a long-term response or resistance to ALK-TKIs. A collaboration is in development to confirm the validity of this protein signature in a larger cohort, and with next-generation ALK-TKIs.
6f8b794f3246b0c1e1780bb4d4d5dc53
-
+
P2.09 - Pathology (Not CME Accredited Session) (ID 958)
- Event: WCLC 2018
- Type: Poster Viewing in the Exhibit Hall
- Track:
- Presentations: 1
- Moderators:
- Coordinates: 9/25/2018, 16:45 - 18:00, Exhibit Hall
-
+
P2.09-07 - Does Metastatic Site Matter for PD-L1 Testing in Stage IV NSCLC? (ID 13385)
16:45 - 18:00 | Author(s): Victor Cohen
- Abstract
Background
Stage IV non-small cell lung cancer (NSCLC) often presents with metastasis to multiple distant sites. Currently PD-L1 expression by immunohistochemistry (IHC) testing with Tumor Proportion Score (TPS) ≥ 50% and ≥1% is required for first- and second-line Pembrolizumab treatment respectively. However, it is not well known if PD-L1 expression differs in NSCLC specimens sampled from different distant metastatic sites. In this study, we evaluate PD-L1 expression in distant metastatic sites.
a9ded1e5ce5d75814730bb4caaf49419 Method
A total of 400 NSCLC specimens from distant metastatic sites are included in this study. The metastatic sites include brain, bone, non-regional lymph nodes, serous membranes (pleura, pericardium and peritoneum) and organs outside the chest (liver, adrenal gland, skin, soft tissue). The samples are either cytology cell blocks, small biopsies or surgical resections. IHC was performed using Dako PD-L1 IHC 22C3 pharmDx. A total of 100 viable tumor cells is required for adequacy. TPS≥ 50% and 1-49% are defined as high and low PD-L1 expression respectively.
4c3880bb027f159e801041b1021e88e8 Result
Overall, the rate of TPS >50% ranges from 36-47% in different metastatic organ sites (Table 1). The prevalence of PD-L1 high and low expression is similar for all distant metastatic sites (P=0.91). Brain metastases have a slightly lower rate of high PD-L1 expression but the difference is not statistically significant.
Table 1. PD-L1 expression in different metastatic sites
Metastatic sites
Tumor Proportion Score (TPS)
Total
≥50%
n (%)
1-49%
n (%)
0%
n (%)
n (%)
Brain
13(36%)
9 (25%)
14(39%)
36 (100%)
Bone
21(44%)
11(23%)
16(33%)
48 (100%)
Nonregional lymph nodes
6(40%)
3(20%)
6 (40%)
15 (100%)
Serous membranes
91(40%)
62(27%)
74(33%)
227 (100%)
Organ outside chest
35(47%)
20(27%)
19(26%)
74 (100%)
Total
166(42%)
105(26%)
129(32%)
400 (100%)
P=0.91
8eea62084ca7e541d918e823422bd82e Conclusion
Our results suggest that the specimens for PD-L1 IHC testing can be sampled from any accessible distant metastatic site.
6f8b794f3246b0c1e1780bb4d4d5dc53


